Molecular dynamics analyses of cross-beta-spine steric zipper models: beta-sheet twisting and aggregation
- PMID: 16864786
- PMCID: PMC1544204
- DOI: 10.1073/pnas.0602345103
Molecular dynamics analyses of cross-beta-spine steric zipper models: beta-sheet twisting and aggregation
Abstract
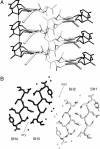
The structural characterization of amyloid fibers is one of the most investigated areas in structural biology. The structural motif, denoted as steric zipper, recently discovered for the peptide GNNQQNY [Nelson, R., Sawaya, M. R., Balbirnie, M., Madsen, A. O., Riekel, C., Grothe, R. & Eisenberg, D. (2005) Nature 435, 773-778], is expected to exert strong influence in this field. To obtain further insights into the features of this unique structural motif, we report several molecular dynamics simulations of various GNNQQNY aggregates. Our analyses show that even pairs of beta-sheets composed of a limited number of beta-strands are stable in the 20-ns time interval considered, which suggests that steric zipper interactions at a beta-sheet-beta-sheet interface strongly contribute to the stability of these aggregates. Moreover, although the basic features of side chain-side chain interactions are preserved in the simulation, the backbone structure undergoes significant variations. Upon equilibration, a significant twist of the beta-strands that compose the beta-sheets is observed. These results demonstrate that the occurrence of steric zipper interactions is compatible with flat and twisted beta-sheets. Molecular dynamics simulations carried out on two pairs of beta-sheets, separated in the crystal state by a hydrated interface, lead to interesting results. The two pairs of sheets, while twisting, associate through stable peptide-peptide interactions. These findings provide insight into the mechanism that leads to the formation of higher aggregates. On these bases, it is possible to reconcile the crystallographic and the EM data on the size of the basic GNNQQNY fiber unit.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

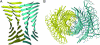

Similar articles
-
Dynamics and stability of amyloid-like steric zipper assemblies with hydrophobic dry interfaces.Biopolymers. 2009 Dec;91(12):1161-71. doi: 10.1002/bip.21182. Biopolymers. 2009. PMID: 19280623
-
Structural stability and dynamics of an amyloid-forming peptide GNNQQNY from the yeast prion sup-35.Biophys J. 2006 Aug 1;91(3):824-33. doi: 10.1529/biophysj.106.083246. Epub 2006 May 5. Biophys J. 2006. PMID: 16679374 Free PMC article.
-
Molecular dynamics simulation study on the molecular structures of the amylin fibril models.J Phys Chem B. 2012 Dec 6;116(48):13991-9. doi: 10.1021/jp308708h. Epub 2012 Nov 26. J Phys Chem B. 2012. PMID: 23145779
-
Understanding amyloid fibril nucleation and aβ oligomer/drug interactions from computer simulations.Acc Chem Res. 2014 Feb 18;47(2):603-11. doi: 10.1021/ar4002075. Epub 2013 Dec 24. Acc Chem Res. 2014. PMID: 24368046 Review.
-
Defining the Landscape of the Pauling-Corey Rippled Sheet: An Orphaned Motif Finding New Homes.Acc Chem Res. 2021 May 18;54(10):2488-2501. doi: 10.1021/acs.accounts.1c00084. Epub 2021 Apr 26. Acc Chem Res. 2021. PMID: 33901396 Free PMC article. Review.
Cited by
-
Structural complexity of a composite amyloid fibril.J Am Chem Soc. 2011 Sep 21;133(37):14686-98. doi: 10.1021/ja203736z. Epub 2011 Aug 23. J Am Chem Soc. 2011. PMID: 21766841 Free PMC article.
-
Dry amyloid fibril assembly in a yeast prion peptide is mediated by long-lived structures containing water wires.Proc Natl Acad Sci U S A. 2010 Dec 14;107(50):21459-64. doi: 10.1073/pnas.1008616107. Epub 2010 Nov 22. Proc Natl Acad Sci U S A. 2010. PMID: 21098298 Free PMC article.
-
Structures and diseases.Nat Struct Mol Biol. 2008 Feb;15(2):117-20. doi: 10.1038/nsmb0208-117. Nat Struct Mol Biol. 2008. PMID: 18250627 Free PMC article.
-
The centriolar protein CPAP G-box: an amyloid fibril in a single domain.Biochem Soc Trans. 2015 Oct;43(5):838-43. doi: 10.1042/BST20150082. Biochem Soc Trans. 2015. PMID: 26517891 Free PMC article. Review.
-
Modeling the Alzheimer Abeta17-42 fibril architecture: tight intermolecular sheet-sheet association and intramolecular hydrated cavities.Biophys J. 2007 Nov 1;93(9):3046-57. doi: 10.1529/biophysj.107.110700. Epub 2007 Aug 3. Biophys J. 2007. PMID: 17675353 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

