Cellular asymmetry and individuality in directional sensing
- PMID: 16864788
- PMCID: PMC1544207
- DOI: 10.1073/pnas.0601909103
Cellular asymmetry and individuality in directional sensing
Abstract
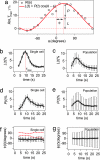
It is generally assumed that single cells in an isogenic population, when exposed to identical environments, exhibit the same behavior. However, it is becoming increasingly clear that, even in a genetically identical population, cellular behavior can vary significantly among cells. Here we explore this variability in the gradient-sensing response of Dictyostelium cells when exposed to repeated spatiotemporal pulses of chemoattractant. Our experiments show the response of a single cell to be highly reproducible from pulse to pulse. In contrast, a large variability in the response direction and magnitude is observed from cell to cell, even when different cells are exposed to the same pulse. First, these results indicate that the gradient-sensing network has inherent asymmetries that can significantly impact the ability of cells to faithfully sense the direction of extracellular signals (cellular asymmetry). Second, we find that the magnitude of this asymmetry varies greatly among cells. Some cells are able to accurately follow the direction of an extracellular stimulus, whereas, in other cells, the intracellular asymmetry dominates, resulting in a polarization axis that is independent of the direction of the extracellular cue (cellular individuality). We integrate these experimental findings into a model that treats the effective signal a cell detects as the product of the extracellular signal and the asymmetric intracellular signal. With this model we successfully predict the population response. This cellular individuality and asymmetry might fundamentally limit the fidelity of signal detection; in contrast, however, it might be beneficial by diversifying phenotypes in isogenic populations.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures



References
-
- Rao C. V., Wolf D. M., Arkin A. P. Nature. 2002;420:231–237. - PubMed
-
- Kaern M., Elston T. C., Blake W. J., Collins J. J. Nat. Rev. Genet. 2005;6:451–464. - PubMed
-
- Spudich J. L., Koshland D. E., Jr Nature. 1976;262:467–471. - PubMed
-
- Korobkova E., Emonet T., Vilar J. M., Shimizu T. S., Cluzel P. Nature. 2004;428:574–578. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

