Analysis of cell-based RNAi screens
- PMID: 16869968
- PMCID: PMC1779553
- DOI: 10.1186/gb-2006-7-7-R66
Analysis of cell-based RNAi screens
Abstract
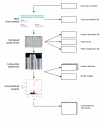
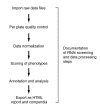
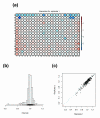
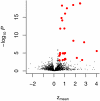
RNA interference (RNAi) screening is a powerful technology for functional characterization of biological pathways. Interpretation of RNAi screens requires computational and statistical analysis techniques. We describe a method that integrates all steps to generate a scored phenotype list from raw data. It is implemented in an open-source Bioconductor/R package, cellHTS (http://www.dkfz.de/signaling/cellHTS). The method is useful for the analysis and documentation of individual RNAi screens. Moreover, it is a prerequisite for the integration of multiple experiments.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

