De novo assembly of genuine replication forks on an immobilized circular plasmid in Xenopus egg extracts
- PMID: 16870720
- PMCID: PMC1540734
- DOI: 10.1093/nar/gkl512
De novo assembly of genuine replication forks on an immobilized circular plasmid in Xenopus egg extracts
Abstract
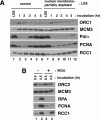
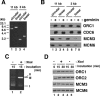
We describe an improved model of DNA replication in Xenopus egg extracts, in which a circular plasmid immobilized on paramagnetic beads is used as a template. DNA synthesis occurred on either circular or linear plasmids coupled to the beads, but only DNA synthesis on the circular plasmid was inhibited by geminin and a CDK inhibitor, p21. DNA synthesis on the circular plasmid occurred after a time lag, during which nuclear formation was probably occurring. Although pre-replicative complexes (pre-RCs) were formed soon after mixing plasmids with egg extracts, binding of CDC45, RPA, Pol alpha, delta and epsilon, and PCNA to the circular plasmid was delayed, but still correlated with DNA synthesis. Moreover, p21 inhibited binding of these replication fork proteins to the circular plasmid. Therefore, the circular plasmid, but not the linear plasmid, assembles bona fide replication forks in egg extracts. We conclude that this improved replication system will be useful for studying the mechanism of formation of replication forks in eukaryotic DNA replication.
Figures



Similar articles
-
DNA is a co-factor for its own replication in Xenopus egg extracts.Nucleic Acids Res. 2011 Jan;39(2):545-55. doi: 10.1093/nar/gkq739. Epub 2010 Sep 21. Nucleic Acids Res. 2011. PMID: 20861001 Free PMC article.
-
DNA polymerase epsilon is required for coordinated and efficient chromosomal DNA replication in Xenopus egg extracts.Proc Natl Acad Sci U S A. 2001 Apr 24;98(9):4978-83. doi: 10.1073/pnas.081088798. Epub 2001 Apr 10. Proc Natl Acad Sci U S A. 2001. PMID: 11296256 Free PMC article.
-
Efficient plasmid DNA replication in Xenopus egg extracts does not depend on prior chromatin assembly.J Biol Chem. 1995 Dec 15;270(50):29676-81. doi: 10.1074/jbc.270.50.29676. J Biol Chem. 1995. PMID: 8530355
-
Dynamics of DNA binding of replication initiation proteins during de novo formation of pre-replicative complexes in Xenopus egg extracts.J Biol Chem. 2006 Apr 21;281(16):10926-34. doi: 10.1074/jbc.M600299200. Epub 2006 Feb 22. J Biol Chem. 2006. PMID: 16497662
-
Xenopus cell-free extracts and their contribution to the study of DNA replication and other complex biological processes.Int J Dev Biol. 2016;60(7-8-9):201-207. doi: 10.1387/ijdb.160142jb. Int J Dev Biol. 2016. PMID: 27759151 Review.
Cited by
-
The DNA unwinding element binding protein DUE-B interacts with Cdc45 in preinitiation complex formation.Mol Cell Biol. 2010 Mar;30(6):1495-507. doi: 10.1128/MCB.00710-09. Epub 2010 Jan 11. Mol Cell Biol. 2010. PMID: 20065034 Free PMC article.
-
Detection of Schistosoma mansoni eggs in feces through their interaction with paramagnetic beads in a magnetic field.PLoS Negl Trop Dis. 2007 Nov 14;1(2):e73. doi: 10.1371/journal.pntd.0000073. PLoS Negl Trop Dis. 2007. PMID: 18060086 Free PMC article.
-
DNA structure-induced recruitment and activation of the Fanconi anemia pathway protein FANCD2.Mol Cell Biol. 2007 Jun;27(12):4283-92. doi: 10.1128/MCB.02196-06. Epub 2007 Apr 9. Mol Cell Biol. 2007. PMID: 17420278 Free PMC article.
-
Different roles of the human Orc6 protein in the replication initiation process.Cell Mol Life Sci. 2011 Nov;68(22):3741-56. doi: 10.1007/s00018-011-0675-9. Epub 2011 Apr 2. Cell Mol Life Sci. 2011. PMID: 21461783 Free PMC article.
References
-
- Bell S.P., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. - PubMed
-
- Stillman B. Origin recognition and the chromosome cycle. FEBS Lett. 2005;579:877–884. - PubMed
-
- Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 1997;272:24508–24513. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

