Retraction of synapses and dendritic spines induced by off-target effects of RNA interference
- PMID: 16870727
- PMCID: PMC6674211
- DOI: 10.1523/JNEUROSCI.1957-06.2006
Retraction of synapses and dendritic spines induced by off-target effects of RNA interference
Abstract
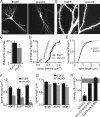
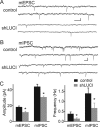
RNA interference (RNAi), which allows selective gene silencing, has been proposed for functional genomic analysis and for the treatment of human disease. However, induction of RNAi in mammalian cells by expression of double-stranded RNA can activate innate antiviral response pathways that perturb off-target gene expression. The activation and functional consequences of these effects in neurons are unknown. We find that expression of subsets of short hairpin RNAs (shRNAs) in rat hippocampal pyramidal neurons can have off-target effects that reduce the complexity of dendritic arbors and trigger the loss of dendritic spines. Morphological changes are accompanied by electrophysiological perturbations in passive membrane properties and a decrease in the number and strength of excitatory and inhibitory synapses. These perturbations depend on the shRNA sequence and are independent of the identity of the targeted protein. Our results indicate that off-target effects of RNAi severely perturb neuronal structure and function and may lead to the functional withdrawal of affected cells from the brain circuitry.
Figures

References
-
- Asano A, Jin HK, Watanabe T (2003). Mouse Mx2 gene: organization, mRNA expression and the role of the interferon-response promoter in its regulation. Gene 306:105–113. - PubMed
-
- Banker G, Goslin K (1998). In: Culturing nerve cells Ed 2 Cambridge, MA: MIT.
-
- Bertram MJ, Berube NG, Hang-Swanson X, Ran Q, Leung JK, Bryce S, Spurgers K, Bick RJ, Baldini A, Ning Y, Clark LJ, Parkinson EK, Barrett JC, Smith JR, Pereira-Smith OM (1999). Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol Cell Biol 19:1479–1485. - PMC - PubMed
-
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, Marshall WS, Khvorova A (2006). 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods 3:199–204. - PubMed
-
- Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R (2003). Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet 34:263–264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
