Comparison and evaluation of methods for generating differentially expressed gene lists from microarray data
- PMID: 16872483
- PMCID: PMC1544358
- DOI: 10.1186/1471-2105-7-359
Comparison and evaluation of methods for generating differentially expressed gene lists from microarray data
Abstract
Background: Numerous feature selection methods have been applied to the identification of differentially expressed genes in microarray data. These include simple fold change, classical t-statistic and moderated t-statistics. Even though these methods return gene lists that are often dissimilar, few direct comparisons of these exist. We present an empirical study in which we compare some of the most commonly used feature selection methods. We apply these to 9 publicly available datasets, and compare, both the gene lists produced and how these perform in class prediction of test datasets.
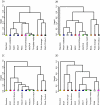
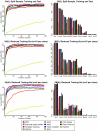
Results: In this study, we compared the efficiency of the feature selection methods; significance analysis of microarrays (SAM), analysis of variance (ANOVA), empirical bayes t-statistic, template matching, maxT, between group analysis (BGA), Area under the receiver operating characteristic (ROC) curve, the Welch t-statistic, fold change, rank products, and sets of randomly selected genes. In each case these methods were applied to 9 different binary (two class) microarray datasets. Firstly we found little agreement in gene lists produced by the different methods. Only 8 to 21% of genes were in common across all 10 feature selection methods. Secondly, we evaluated the class prediction efficiency of each gene list in training and test cross-validation using four supervised classifiers.
Conclusion: We report that the choice of feature selection method, the number of genes in the genelist, the number of cases (samples) and the noise in the dataset, substantially influence classification success. Recommendations are made for choice of feature selection. Area under a ROC curve performed well with datasets that had low levels of noise and large sample size. Rank products performs well when datasets had low numbers of samples or high levels of noise. The Empirical bayes t-statistic performed well across a range of sample sizes.
Figures

References
-
- Long AD, Mangalam HJ, Chan BY, Tolleri L, Hatfield GW, Baldi P. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J Biol Chem. 2001;276:19937–19944. doi: 10.1074/jbc.M010192200. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

