Conservation of alternative polyadenylation patterns in mammalian genes
- PMID: 16872498
- PMCID: PMC1550727
- DOI: 10.1186/1471-2164-7-189
Conservation of alternative polyadenylation patterns in mammalian genes
Abstract
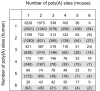
Background: Alternative polyadenylation is a widespread mechanism contributing to transcript diversity in eukaryotes. Over half of mammalian genes are alternatively polyadenylated. Our understanding of poly(A) site evolution is limited by the lack of a reliable identification of conserved, equivalent poly(A) sites among species. We introduce here a working definition of conserved poly(A) sites as sites that are both (i) properly aligned in human and mouse orthologous 3' untranslated regions (UTRs) and (ii) supported by EST or cDNA data in both species.
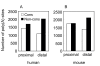
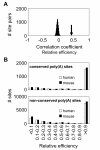
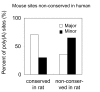
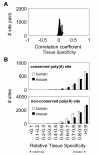
Results: We identified about 4800 such conserved poly(A) sites covering one third of the orthologous gene set studied. Characteristics of conserved poly(A) sites such as processing efficiency and tissue-specificity were analyzed. Conserved sites show a higher processing efficiency but no difference in tissular distribution when compared to non-conserved sites. In general, alternative poly(A) sites are species-specific and involve minor, non-conserved sites that are unlikely to play essential roles. However, there are about 500 genes with conserved tandem poly(A) sites. A significant fraction of these conserved tandems display a conserved arrangement of major/minor sites in their 3' UTR, suggesting that these alternative 3' ends may be under selection.
Conclusion: This analysis allows us to identify potential functional alternative poly(A) sites and provides clues on the selective mechanisms at play in the appearance of multiple poly(A) sites and their maintenance in the 3' UTRs of genes.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

