Genomes of Helicobacter pylori from native Peruvians suggest admixture of ancestral and modern lineages and reveal a western type cag-pathogenicity island
- PMID: 16872520
- PMCID: PMC1553449
- DOI: 10.1186/1471-2164-7-191
Genomes of Helicobacter pylori from native Peruvians suggest admixture of ancestral and modern lineages and reveal a western type cag-pathogenicity island
Abstract
Background: Helicobacter pylori is presumed to be co-evolved with its human host and is a highly diverse gastric pathogen at genetic levels. Ancient origins of H. pylori in the New World are still debatable. It is not clear how different waves of human migrations in South America contributed to the evolution of strain diversity of H. pylori. The objective of our 'phylogeographic' study was to gain fresh insights into these issues through mapping genetic origins of H. pylori of native Peruvians (of Amerindian ancestry) and their genomic comparison with isolates from Spain, and Japan.
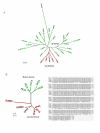
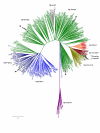
Results: For this purpose, we attempted to dissect genetic identity of strains by fluorescent amplified fragment length polymorphism (FAFLP) analysis, multilocus sequence typing (MLST) of the 7 housekeeping genes (atpA, efp, ureI, ppa, mutY, trpC, yphC) and the sequence analyses of the babB adhesin and oipA genes. The whole cag pathogenicity-island (cagPAI) from these strains was analyzed using PCR and the geographic type of cagA phosphorylation motif EPIYA was determined by gene sequencing. We observed that while European genotype (hp-Europe) predominates in native Peruvian strains, approximately 20% of these strains represent a sub-population with an Amerindian ancestry (hsp-Amerind). All of these strains however, irrespective of their ancestral affiliation harbored a complete, 'western' type cagPAI and the motifs surrounding it. This indicates a possible acquisition of cagPAI by the hsp-Amerind strains from the European strains, during decades of co-colonization.
Conclusion: Our observations suggest presence of ancestral H. pylori (hsp-Amerind) in Peruvian Amerindians which possibly managed to survive and compete against the Spanish strains that arrived to the New World about 500 years ago. We suggest that this might have happened after native Peruvian H. pylori strains acquired cagPAI sequences, either by new acquisition in cag-negative strains or by recombination in cag positive Amerindian strains.
Figures

References
-
- Alm RA, Ling LSL, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, Carmel G, Tummino PJ, Caruso A, Nickelsen MU, Mills DM, Ives C, Gibson R, Merberg D, Mills SD, Jiang Q, Taylor DE, Vovis GF, Trust TJ. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

