Hydrogen tunnelling in enzyme-catalysed H-transfer reactions: flavoprotein and quinoprotein systems
- PMID: 16873125
- PMCID: PMC1647315
- DOI: 10.1098/rstb.2006.1878
Hydrogen tunnelling in enzyme-catalysed H-transfer reactions: flavoprotein and quinoprotein systems
Abstract
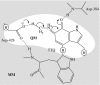
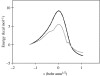
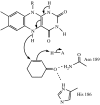
It is now widely accepted that enzyme-catalysed C-H bond breakage occurs by quantum mechanical tunnelling. This paradigm shift in the conceptual framework for these reactions away from semi-classical transition state theory (TST, i.e. including zero-point energy, but with no tunnelling correction) has been driven over the recent years by experimental studies of the temperature dependence of kinetic isotope effects (KIEs) for these reactions in a range of enzymes, including the tryptophan tryptophylquinone-dependent enzymes such as methylamine dehydrogenase and aromatic amine dehydrogenase, and the flavoenzymes such as morphinone reductase and pentaerythritol tetranitrate reductase, which produced observations that are also inconsistent with the simple Bell-correction model of tunnelling. However, these data-especially, the strong temperature dependence of reaction rates and the variable temperature dependence of KIEs-are consistent with other tunnelling models (termed full tunnelling models), in which protein and/or substrate fluctuations generate a configuration compatible with tunnelling. These models accommodate substrate/protein (environment) fluctuations required to attain a configuration with degenerate nuclear quantum states and, when necessary, motion required to increase the probability of tunnelling in these states. Furthermore, tunnelling mechanisms in enzymes are supported by atomistic computational studies performed within the framework of modern TST, which incorporates quantum nuclear effects.
Figures


References
-
- Agarwal P.K, Billeter S.R, Hammes-Schiffer S. Nuclear quantum effects and enzyme dynamics in dihydrofolate reductase catalysis. J. Phys. Chem. B. 2002a;106:3283–3293. doi:10.1021/jp020190v - DOI
-
- Agarwal P.K, Billeter S.R, Rajagopalan P.T, Benkovic S.J, Hammes-Schiffer S. Network of coupled promoting motions in enzyme catalysis. Proc. Natl Acad. Sci. USA. 2002b;99:2794–2799. doi:10.1073/pnas.052005999 - DOI - PMC - PubMed
-
- Alhambra C, Corchado J, Sanchez M, Gao J, Truhlar D. Quantum dynamics of hydride transfer in enzyme catalysis. J. Am. Chem. Soc. 2000;122:8197–8203. doi:10.1021/ja001476l - DOI
-
- Alhambra C, Sanchez M.L, Corchado J, Gao J.L, Truhlar D.G. Quantum mechanical tunneling in methylamine dehydrogenase. Chem. Phys. Lett. 2001;347:512–518. doi:10.1016/S0009-2614(01)00921-6 - DOI
-
- Alhambra C, Sanchez M.L, Corchado J, Gao J, Truhlar D.G. Erratum to: “Quantum mechanical tunneling in methylamine dehydrogenase”. Chem. Phys. Lett. 2002;355:388–394. doi:10.1016/S0009-2614(02)00057-X - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
