Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells
- PMID: 16873239
- PMCID: PMC1563789
- DOI: 10.1128/JVI.00532-06
Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells
Abstract
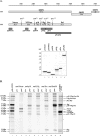
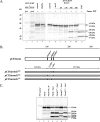
Murine norovirus (MNV) is presently the only member of the genus Norovirus in the Caliciviridae that can be propagated in cell culture. The goal of this study was to elucidate the proteolytic processing strategy of MNV during an authentic replication cycle in cells. A proteolytic cleavage map of the ORF1 polyprotein was generated, and the virus-encoded 3C-like (3CL) proteinase (Pro) mediated cleavage at five dipeptide cleavage sites, 341E/G342, Q705/N706, 870E/G871, 994E/A995, and 1177Q/G1178, that defined the borders of six proteins with the gene order p38.3 (Nterm)-p39.6 (NTPase)-p18.6-p14.3 (VPg)-p19.2 (Pro)-p57.5 (Pol). Bacterially expressed MNV 3CL Pro was sufficient to mediate trans cleavage of the ORF1 polyprotein containing the mutagenized Pro sequence into products identical to those observed during cotranslational processing of the authentic ORF1 polyprotein in vitro and to those observed in MNV-infected cells. Immunoprecipitation and Western blot analysis of proteins produced in virus-infected cells demonstrated efficient cleavage of the proteinase-polymerase precursor. Evidence for additional processing of the Nterm protein in MNV-infected cells by caspase 3 was obtained, and Nterm sequences 118DRPD121 and 128DAMD131 were mapped as caspase 3 cleavage sites by site-directed mutagenesis. The availability of the MNV nonstructural polyprotein cleavage map in concert with a permissive cell culture system should facilitate studies of norovirus replication.
Figures
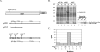




References
-
- Al-Molawi, N., V. A. Beardmore, M. J. Carter, G. E. Kass, and L. O. Roberts. 2003. Caspase-mediated cleavage of the feline calicivirus capsid protein. J Gen. Virol. 84:1237-1244. - PubMed
-
- Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181(Suppl. 2):S336-S348. - PubMed
-
- Belliot, G., S. V. Sosnovtsev, T. Mitra, C. Hammer, M. Garfield, and K. Y. Green. 2003. In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells. J. Virol. 77:10957-10974. - PMC - PubMed
-
- Blakeney, S. J., A. Cahill, and P. A. Reilly. 2003. Processing of Norwalk virus nonstructural proteins by a 3C-like cysteine proteinase. Virology 308:216-224. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

