Prion strain-dependent differences in conversion of mutant prion proteins in cell culture
- PMID: 16873242
- PMCID: PMC1563786
- DOI: 10.1128/JVI.00424-06
Prion strain-dependent differences in conversion of mutant prion proteins in cell culture
Abstract
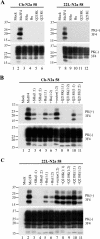
Although the protein-only hypothesis proposes that it is the conformation of abnormal prion protein (PrP(Sc)) that determines strain diversity, the molecular basis of strains remains to be elucidated. In the present study, we generated a series of mutations in the normal prion protein (PrP(C)) in which a single glutamine residue was replaced with a basic amino acid and compared their abilities to convert to PrP(Sc) in cultured neuronal N2a58 cells infected with either the Chandler or 22L mouse-adapted scrapie strain. In mice, these strains generate PrP(Sc) of the same sequence but different conformations, as judged by infrared spectroscopy. Substitutions at codons 97, 167, 171, and 216 generated PrP(C) that resisted conversion and inhibited the conversion of coexpressed wild-type PrP in both Chandler-infected and 22L-infected cells. Interestingly, substitutions at codons 185 and 218 gave strain-dependent effects. The Q185R and Q185K PrP were efficiently converted to PrP(Sc) in Chandler-infected but not 22L-infected cells. Conversely, Q218R and Q218H PrP were converted only in 22L-infected cells. Moreover, the Q218K PrP exerted a potent inhibitory effect on the conversion of coexpressed wild-type PrP in Chandler-infected cells but had little effect on 22L-infected cells. These results show that two strains with the same PrP sequence but different conformations have differing abilities to convert the same mutated PrP(C).
Figures



Similar articles
-
Disulfide-crosslink scanning reveals prion-induced conformational changes and prion strain-specific structures of the pathological prion protein PrPSc.J Biol Chem. 2018 Aug 17;293(33):12730-12740. doi: 10.1074/jbc.RA117.001633. Epub 2018 Jun 22. J Biol Chem. 2018. PMID: 29934306 Free PMC article.
-
Conformational properties of prion strains can be transmitted to recombinant prion protein fibrils in real-time quaking-induced conversion.J Virol. 2014 Oct;88(20):11791-801. doi: 10.1128/JVI.00585-14. Epub 2014 Jul 30. J Virol. 2014. PMID: 25078700 Free PMC article.
-
Inhibition of prion propagation in scrapie-infected mouse neuroblastoma cell lines using mouse monoclonal antibodies against prion protein.Biochem Biophys Res Commun. 2005 Sep 16;335(1):197-204. doi: 10.1016/j.bbrc.2005.07.063. Biochem Biophys Res Commun. 2005. PMID: 16061207
-
Prion encephalopathies of animals and humans.Dev Biol Stand. 1993;80:31-44. Dev Biol Stand. 1993. PMID: 8270114 Review.
-
Molecular biology and transgenetics of prion diseases.Crit Rev Biochem Mol Biol. 1991;26(5-6):397-438. doi: 10.3109/10409239109086789. Crit Rev Biochem Mol Biol. 1991. PMID: 1684745 Review.
Cited by
-
Prions disturb post-Golgi trafficking of membrane proteins.Nat Commun. 2013;4:1846. doi: 10.1038/ncomms2873. Nat Commun. 2013. PMID: 23673631
-
A Single Amino Acid Substitution, Found in Mammals with Low Susceptibility to Prion Diseases, Delays Propagation of Two Prion Strains in Highly Susceptible Transgenic Mouse Models.Mol Neurobiol. 2019 Sep;56(9):6501-6511. doi: 10.1007/s12035-019-1535-0. Epub 2019 Mar 7. Mol Neurobiol. 2019. PMID: 30847740
-
Chaperone-mediated disaggregation of infectious prions releases particles that seed new prion formation in a strain-specific manner.J Biol Chem. 2025 Jan;301(1):108062. doi: 10.1016/j.jbc.2024.108062. Epub 2024 Dec 9. J Biol Chem. 2025. PMID: 39662829 Free PMC article.
-
Effect of hydrophobic mutations in the H2-H3 subdomain of prion protein on stability and conversion in vitro and in vivo.PLoS One. 2011;6(9):e24238. doi: 10.1371/journal.pone.0024238. Epub 2011 Sep 1. PLoS One. 2011. PMID: 21909425 Free PMC article.
-
A proposed mechanism for the promotion of prion conversion involving a strictly conserved tyrosine residue in the β2-α2 loop of PrPC.J Biol Chem. 2014 Apr 11;289(15):10660-10667. doi: 10.1074/jbc.M114.549030. Epub 2014 Mar 4. J Biol Chem. 2014. PMID: 24596090 Free PMC article.
References
-
- Arima, K., N. Nishida, S. Sakaguchi, K. Shigematsu, R. Atarashi, N. Yamaguchi, D. Yoshikawa, J. Yoon, K. Watanabe, N. Kobayashi, S. Mouillet-Richard, S. Lehmann, and S. Katamine. 2005. Biological and biochemical characteristics of prion strains conserved in persistently infected cell cultures. J. Virol. 79:7104-7112. - PMC - PubMed
-
- Atarashi, R., N. Nishida, K. Shigematsu, S. Goto, T. Kondo, S. Sakaguchi, and S. Katamine. 2003. Deletion of N-terminal residues 23-88 from prion protein (PrP) abrogates the potential to rescue PrP-deficient mice from PrP-like protein/Doppel-induced neurodegeneration. J. Biol. Chem. 278:28944-28949. - PubMed
-
- Belt, P. B., I. H. Muileman, B. E. Schreuder, R. Bos-de Ruijter, A. L. Gielkens, and M. A. Smits. 1995. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. J. Gen. Virol. 76:509-517. - PubMed
-
- Ben-Zaken, O., S. Tzaban, Y. Tal, L. Horonchik, J. D. Esko, I. Vlodavsky, and A. Taraboulos. 2003. Cellular heparan sulfate participates in the metabolism of prions. J. Biol. Chem. 278:40041-40049. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

