Mutation of dileucine-like motifs in the human immunodeficiency virus type 1 capsid disrupts virus assembly, gag-gag interactions, gag-membrane binding, and virion maturation
- PMID: 16873251
- PMCID: PMC1563813
- DOI: 10.1128/JVI.00355-06
Mutation of dileucine-like motifs in the human immunodeficiency virus type 1 capsid disrupts virus assembly, gag-gag interactions, gag-membrane binding, and virion maturation
Abstract
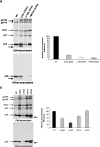
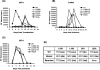
The human immunodeficiency virus type 1 (HIV-1) Gag precursor protein Pr55(Gag) drives the assembly and release of virus-like particles in the infected cell. The capsid (CA) domain of Gag plays an important role in these processes by promoting Gag-Gag interactions during assembly. The C-terminal domain (CTD) of CA contains two dileucine-like motifs (L189/L190 and I201/L202) implicated in regulating the localization of Gag to multivesicular bodies (MVBs). These dileucine-like motifs are located in the vicinity of the CTD dimer interface, a region of CA critical for Gag-Gag interactions during virus assembly and CA-CA interactions during core formation. To study the importance of the CA dileucine-like motifs in various aspects of HIV-1 replication, we introduced a series of mutations into these motifs in the context of a full-length, infectious HIV-1 molecular clone. CA mutants LL189,190AA and IL201,202AA were both severely impaired in virus particle production because of a variety of defects in the binding of Gag to membrane, Gag multimerization, and CA folding. In contrast to the model suggesting that the CA dileucine-like motifs regulate MVB targeting, the IL201,202AA mutation did not alter Gag localization to the MVB in either HeLa cells or macrophages. Revertants of single-amino-acid substitution mutants were obtained that no longer contained dileucine-like motifs but were nevertheless fully replication competent. The varied phenotypes of the mutants reported here provide novel insights into the interplay among Gag multimerization, membrane binding, virus assembly, CA dimerization, particle maturation, and virion infectivity.
Figures




References
-
- Bonifacino, J. S., and L. M. Traub. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395-447. - PubMed
-
- Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

