Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections
- PMID: 16873257
- PMCID: PMC1563799
- DOI: 10.1128/JVI.00743-06
Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections
Abstract
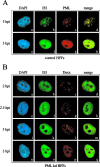
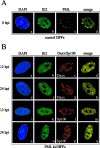
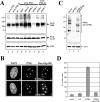
Several viruses, including human cytomegalovirus (HCMV), encode proteins that colocalize with a cellular subnuclear structure known as ND10. Since only viral DNA deposited at ND10 initiates transcription, ND10 structures were hypothesized to be essential for viral replication. On the other hand, interferon treatment induces an up-regulation of ND10 structures and viruses have evolved polypeptides that disperse the dot-like accumulation of ND10 proteins, suggesting that ND10 could also be part of an intrinsic defense mechanism. In order to obtain evidence for either a proviral or an antiviral function of ND10, we generated primary human fibroblasts with a stable, short interfering RNA-mediated knockdown (kd) of PML. In these cells, other ND10-associated proteins like hDaxx showed a diffuse nuclear distribution. Interestingly, we observed that HCMV infection induced the de novo formation of ND10-like hDaxx and Sp100 accumulations that colocalized with IE2 and were disrupted, in the apparent absence of PML, in an IE1-dependent manner during the first hours after infection. Furthermore, infection of PML-kd cells with wild-type HCMV at a low multiplicity of infection resulted in enhanced replication. In particular, a significantly increased plaque formation was detected, suggesting that more cells are able to support initiation of replication in the absence of PML. While there was no difference in viral DNA uptake between PML-kd and control cells, we observed a considerable increase in the number of immediate-early (IE) protein-positive cells, indicating that the depletion of PML augments the initiation of viral IE gene expression. These results strongly suggest that PML functions as part of an intrinsic immune mechanism against cytomegalovirus infections.
Figures



Similar articles
-
Recruitment of human cytomegalovirus immediate-early 2 protein onto parental viral genomes in association with ND10 in live-infected cells.J Virol. 2007 Sep;81(18):10123-36. doi: 10.1128/JVI.01009-07. Epub 2007 Jul 11. J Virol. 2007. PMID: 17626080 Free PMC article.
-
Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection.Virology. 2000 Aug 15;274(1):39-55. doi: 10.1006/viro.2000.0448. Virology. 2000. PMID: 10936087
-
Contribution of the Major ND10 Proteins PML, hDaxx and Sp100 to the Regulation of Human Cytomegalovirus Latency and Lytic Replication in the Monocytic Cell Line THP-1.Viruses. 2015 Jun 5;7(6):2884-907. doi: 10.3390/v7062751. Viruses. 2015. PMID: 26057166 Free PMC article.
-
Intrinsic cellular defense mechanisms targeting human cytomegalovirus.Virus Res. 2011 May;157(2):128-33. doi: 10.1016/j.virusres.2010.10.002. Epub 2010 Oct 8. Virus Res. 2011. PMID: 20934469 Review.
-
Review: properties and assembly mechanisms of ND10, PML bodies, or PODs.J Struct Biol. 2000 Apr;129(2-3):278-87. doi: 10.1006/jsbi.2000.4239. J Struct Biol. 2000. PMID: 10806078 Review.
Cited by
-
Intrinsic host restriction factors of human cytomegalovirus replication and mechanisms of viral escape.World J Virol. 2016 Aug 12;5(3):87-96. doi: 10.5501/wjv.v5.i3.87. World J Virol. 2016. PMID: 27563536 Free PMC article. Review.
-
Nuclear domain 10-associated proteins recognize and segregate intranuclear DNA/protein complexes to negate gene expression.Virol J. 2012 Sep 28;9:222. doi: 10.1186/1743-422X-9-222. Virol J. 2012. PMID: 23021128 Free PMC article.
-
Specification of the NF-kappaB transcriptional response by p65 phosphorylation and TNF-induced nuclear translocation of IKK epsilon.Nucleic Acids Res. 2010 Oct;38(18):6029-44. doi: 10.1093/nar/gkq439. Epub 2010 May 27. Nucleic Acids Res. 2010. PMID: 20507904 Free PMC article.
-
Tegument protein control of latent herpesvirus establishment and animation.Herpesviridae. 2011 Feb 8;2(1):3. doi: 10.1186/2042-4280-2-3. Herpesviridae. 2011. PMID: 21429246 Free PMC article.
-
Differential functions of interferon-upregulated Sp100 isoforms: herpes simplex virus type 1 promoter-based immediate-early gene suppression and PML protection from ICP0-mediated degradation.J Virol. 2009 May;83(10):5168-80. doi: 10.1128/JVI.02083-08. Epub 2009 Mar 11. J Virol. 2009. PMID: 19279115 Free PMC article.
References
-
- Ahn, J. H., E. J. Brignole III, and G. S. Hayward. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol. 18:4899-4913. - PMC - PubMed
-
- Ahn, J. H., and G. S. Hayward. 2000. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology 274:39-55. - PubMed
-
- Ahn, J. H., W. J. Jang, and G. S. Hayward. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J. Virol. 73:10458-10471. - PMC - PubMed
-
- Andreoni, M., M. Faircloth, L. Vugler, and W. J. Britt. 1989. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J. Virol. Methods 23:157-167. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

