Requirement of the N-terminal region of orthobunyavirus nonstructural protein NSm for virus assembly and morphogenesis
- PMID: 16873265
- PMCID: PMC1563826
- DOI: 10.1128/JVI.00579-06
Requirement of the N-terminal region of orthobunyavirus nonstructural protein NSm for virus assembly and morphogenesis
Abstract
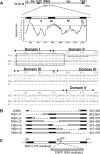
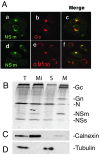
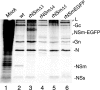
The nonstructural protein NSm of Bunyamwera virus (BUNV), the prototype of the Bunyaviridae family, is encoded by the M segment in a polyprotein precursor, along with the virion glycoproteins, in the order Gn-NSm-Gc. As little is known of its function, we examined the intracellular localization, membrane integrality, and topology of NSm and its role in virus replication. We confirmed that NSm is an integral membrane protein and that it localizes in the Golgi complex, together with Gn and Gc. Coimmunoprecipitation assays and yeast two-hybrid analysis demonstrated that NSm was able to interact with other viral proteins. NSm is predicted to contain three hydrophobic (I, III, and V) and two nonhydrophobic (II and IV) domains. The N-terminal nonhydrophobic domain II was found in the lumen of an intracellular compartment. A novel BUNV assembly assay was developed to monitor the formation of infectious virus-like-particles (VLPs). Using this assay, we showed that deletions of either the complete NSm coding region or domains I, II, and V individually seriously compromised VLP production. Consistently, we were unable to rescue viable viruses by reverse genetics from cDNA constructs that contained the same deletions. However, we could generate mutant BUNV with deletions in NSm domains III and IV and also a recombinant virus with the green fluorescent protein open reading frame inserted into NSm domain IV. The mutant viruses displayed differences in their growth properties. Overall, our data showed that the N-terminal region of NSm, which includes domain I and part of domain II, is required for virus assembly and that the C-terminal hydrophobic domain V may function as an internal signal sequence for the Gc glycoprotein.
Figures



Similar articles
-
Role of the cytoplasmic tail domains of Bunyamwera orthobunyavirus glycoproteins Gn and Gc in virus assembly and morphogenesis.J Virol. 2007 Sep;81(18):10151-60. doi: 10.1128/JVI.00573-07. Epub 2007 Jul 3. J Virol. 2007. PMID: 17609275 Free PMC article.
-
Bunyamwera orthobunyavirus glycoprotein precursor is processed by cellular signal peptidase and signal peptide peptidase.Proc Natl Acad Sci U S A. 2016 Aug 2;113(31):8825-30. doi: 10.1073/pnas.1603364113. Epub 2016 Jul 20. Proc Natl Acad Sci U S A. 2016. PMID: 27439867 Free PMC article.
-
Characterization of Maguari orthobunyavirus mutants suggests the nonstructural protein NSm is not essential for growth in tissue culture.Virology. 2006 Apr 25;348(1):224-32. doi: 10.1016/j.virol.2005.12.026. Epub 2006 Jan 30. Virology. 2006. PMID: 16445958
-
Visualizing the replication cycle of bunyamwera orthobunyavirus expressing fluorescent protein-tagged Gc glycoprotein.J Virol. 2010 Sep;84(17):8460-9. doi: 10.1128/JVI.00902-10. Epub 2010 Jun 23. J Virol. 2010. PMID: 20573824 Free PMC article.
-
Mechanisms of bunyavirus morphogenesis and egress.J Gen Virol. 2023 Apr;104(4). doi: 10.1099/jgv.0.001845. J Gen Virol. 2023. PMID: 37083579 Review.
Cited by
-
Two Akabane virus glycoprotein Gc domains induce neutralizing antibodies in mice.J Vet Med Sci. 2022 Apr 13;84(4):538-542. doi: 10.1292/jvms.21-0670. Epub 2022 Feb 23. J Vet Med Sci. 2022. PMID: 35197396 Free PMC article.
-
A positive-sense single-stranded RNA virus acquired a negative-sense open reading frame through recombination.PLoS Pathog. 2025 Apr 8;21(4):e1013015. doi: 10.1371/journal.ppat.1013015. eCollection 2025 Apr. PLoS Pathog. 2025. PMID: 40198687 Free PMC article.
-
Minigenomes, transcription and replication competent virus-like particles and beyond: reverse genetics systems for filoviruses and other negative stranded hemorrhagic fever viruses.Antiviral Res. 2011 Aug;91(2):195-208. doi: 10.1016/j.antiviral.2011.06.003. Epub 2011 Jun 14. Antiviral Res. 2011. PMID: 21699921 Free PMC article. Review.
-
Systems to establish bunyavirus genome replication in the absence of transcription.J Virol. 2013 Jul;87(14):8205-12. doi: 10.1128/JVI.00371-13. Epub 2013 May 22. J Virol. 2013. PMID: 23698297 Free PMC article.
-
Recent Advances in Bunyavirus Reverse Genetics Research: Systems Development, Applications, and Future Perspectives.Front Microbiol. 2021 Dec 7;12:771934. doi: 10.3389/fmicb.2021.771934. eCollection 2021. Front Microbiol. 2021. PMID: 34950119 Free PMC article. Review.
References
-
- Bishop, D. H. 1996. Biology and molecular biology of bunyaviruses, p. 19-61. In R. M. Elliott (ed.), The Bunyaviridae. Plenum Press, New York, N.Y.
-
- Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. - PMC - PubMed
-
- Bupp, K., K. Stillmock, and F. Gonzalez-Scarano. 1996. Analysis of the intracellular transport properties of recombinant La Crosse virus glycoproteins. Virology 220:485-490. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

