The self primer of the long terminal repeat retrotransposon Tf1 is not removed during reverse transcription
- PMID: 16873283
- PMCID: PMC1563812
- DOI: 10.1128/JVI.01915-05
The self primer of the long terminal repeat retrotransposon Tf1 is not removed during reverse transcription
Abstract
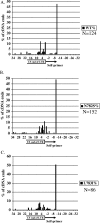
The long terminal repeat retrotransposon Tf1 of Schizosaccharomyces pombe uses a unique mechanism of self priming to initiate reverse transcription. Instead of using a tRNA, Tf1 primes minus-strand synthesis with an 11-nucleotide RNA removed from the 5' end of its own transcript. We tested whether the self primer of Tf1 was similar to tRNA primers in being removed from the cDNA by RNase H. Our analysis of Tf1 cDNA extracted from virus-like particles revealed the surprising observation that the dominant species of cDNA retained the self primer. This suggests that integration of the cDNA relies on mechanisms other than reverse transcription to remove the primer.
Figures

Similar articles
-
The Long Terminal Repeat Retrotransposons Tf1 and Tf2 of Schizosaccharomyces pombe.Microbiol Spectr. 2015 Aug;3(4):10.1128/microbiolspec.MDNA3-0040-2014. doi: 10.1128/microbiolspec.MDNA3-0040-2014. Microbiol Spectr. 2015. PMID: 26350316 Free PMC article. Review.
-
The removal of RNA primers from DNA synthesized by the reverse transcriptase of the retrotransposon Tf1 is stimulated by Tf1 integrase.J Virol. 2012 Jun;86(11):6222-30. doi: 10.1128/JVI.00009-12. Epub 2012 Apr 4. J Virol. 2012. PMID: 22491446 Free PMC article.
-
The reverse transcriptase of the Tf1 retrotransposon has a specific novel activity for generating the RNA self-primer that is functional in cDNA synthesis.J Virol. 2008 Nov;82(21):10906-10. doi: 10.1128/JVI.01370-08. Epub 2008 Aug 27. J Virol. 2008. PMID: 18753200 Free PMC article.
-
Evidence for the packaging of multiple copies of Tf1 mRNA into particles and the trans priming of reverse transcription.J Virol. 2000 Aug;74(15):7164-70. doi: 10.1128/jvi.74.15.7164-7170.2000. J Virol. 2000. PMID: 10888658 Free PMC article.
-
"In the beginning": initiation of minus strand DNA synthesis in retroviruses and LTR-containing retrotransposons.Biochemistry. 2003 Dec 16;42(49):14349-55. doi: 10.1021/bi030201q. Biochemistry. 2003. PMID: 14661945 Review.
Cited by
-
The diversity of retrotransposons and the properties of their reverse transcriptases.Virus Res. 2008 Jun;134(1-2):221-34. doi: 10.1016/j.virusres.2007.12.010. Epub 2008 Feb 7. Virus Res. 2008. PMID: 18261821 Free PMC article. Review.
-
Single-Nucleotide-Specific Targeting of the Tf1 Retrotransposon Promoted by the DNA-Binding Protein Sap1 of Schizosaccharomyces pombe.Genetics. 2015 Nov;201(3):905-24. doi: 10.1534/genetics.115.181602. Epub 2015 Sep 9. Genetics. 2015. PMID: 26358720 Free PMC article.
-
The Long Terminal Repeat Retrotransposons Tf1 and Tf2 of Schizosaccharomyces pombe.Microbiol Spectr. 2015 Aug;3(4):10.1128/microbiolspec.MDNA3-0040-2014. doi: 10.1128/microbiolspec.MDNA3-0040-2014. Microbiol Spectr. 2015. PMID: 26350316 Free PMC article. Review.
-
Identification of an integrase-independent pathway of retrotransposition.Sci Adv. 2022 Jul;8(26):eabm9390. doi: 10.1126/sciadv.abm9390. Epub 2022 Jun 29. Sci Adv. 2022. PMID: 35767609 Free PMC article.
-
The removal of RNA primers from DNA synthesized by the reverse transcriptase of the retrotransposon Tf1 is stimulated by Tf1 integrase.J Virol. 2012 Jun;86(11):6222-30. doi: 10.1128/JVI.00009-12. Epub 2012 Apr 4. J Virol. 2012. PMID: 22491446 Free PMC article.
References
-
- Butler, M., T. Goodwin, M. Simpson, M. Singh, and R. Poulter. 2001. Vertebrate LTR retrotransposons of the Tf1/Sushi group. J. Mol. Evol. 52:260-274. - PubMed
-
- Champoux, J. 1993. Roles of ribonuclease H in reverse transcription, p. 103-117. In A. Skulka and S. Goff (ed.), Reverse transcriptase. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

