Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes
- PMID: 16874071
- PMCID: PMC4007489
- DOI: 10.4161/auto.2229
Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes
Abstract
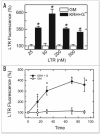
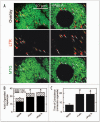
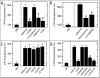
Mitochondria become targets for autophagic degradation after nutrient deprivation, a process also termed mitophagy. In this study, we used LysoTracker Red (LTR) and MitoTracker Green to characterize the kinetics of autophagosomal proliferation and mitophagy in cultured rat hepatocytes. Autophagy induced by nutrient deprivation plus glucagon increased LTR uptake assessed with a fluorescence plate reader and the number of LTR-labeled acidic organelles assessed with confocal microscopy in individual hepatocytes both by 4- to 6-fold. Serial imaging of hepatocytes coloaded with MitoTracker Green (MTG) revealed an average mitochondrial digestion time of 7.5 min after autophagic induction. In the presence of protease inhibitors, digestion time more than doubled, and the total number of LTR-labeled organelles increased about 40%, but the proportion of the LTR-labeled acidic organelles containing MTG fluorescence remained constant at about 75%. Autophagy inhibitors, 3-methyladenine, wortmannin and LY204002, suppressed the increase of LTR uptake after nutrient deprivation by up to 85%, confirming that increased LTR uptake reflected autophagy induction. Cyclosporin A and NIM811, specific inhibitors of the mitochondrial permeability transition (MPT), also decreased LTR uptake, whereas tacrolimus, an immunosuppressive reagent that does not inhibit the MPT, was without effect. In addition, the c-Jun N-terminal kinase (JNK) inhibitors, SCP25041 and SP600125, blocked LTR uptake by 47% and 61%, respectively, but ERK1, p38 and caspase inhibitors had no effect. The results show that mitochondria once selected for mitophagy are rapidly digested and support the concept that mitochondrial autophagy involves the MPT and signaling through PI3 kinase and possibly JNK.
Figures


References
-
- Arstila AU, Shelburne DJ, Trump BF. Studies on cellular autophagocytosis. A histochemical study on sequential alterations of mitochondria in the glucagon-induced autophagy vacuoles of rat liver. Lab Invest. 1972;27:317–23. - PubMed
-
- Lardeux BR, Mortimore GE. Amino acid and hormonal control of macromolecular turnover in perfused rat liver. Evidence for selective autophagy. J Biol Chem. 1987;262:14514–9. - PubMed
-
- Shworer CM, Shiffer KA, Mortimer GE. Quantitative relationship between autophagy and the proteolysis during graded amino acid deprivation in perfused rat liver. J Biol Chem. 1981;256:7652–8. - PubMed
-
- Mortimore GE, Poso AR. Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu Rev Nutr. 1987;5:49–70. - PubMed
-
- Yu QC, Marzella L. Response of autophagic protein degradation to physiologic and pathologic stimuli in rat hepatocyte monolayer cultures. Lab Inves. 1988;58:643–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
