Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b
- PMID: 16874306
- PMCID: PMC1538567
- DOI: 10.1038/sj.emboj.7601244
Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b
Abstract
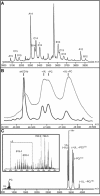
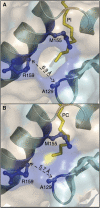
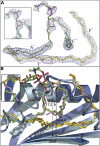
CD1 proteins present lipid antigens to T cells. The antigens are acquired in the endosomal compartments. This raises the question of how the large hydrophobic CD1 pockets are preserved between the moment of biosynthesis in the endoplasmic reticulum and arrival to the endosomes. To address this issue, the natural ligands associated with a soluble form of human CD1b have been investigated. Using isoelectric focusing, native mass spectrometry and resolving the crystal structure at 1.8 A resolution, we found that human CD1b is simultaneously associated with endogenous phosphatidylcholine (PC) and a 41-44 carbon atoms-long spacer molecule. The two lipids appear to work in concert to stabilize the CD1b groove, their combined size slightly exceeding the maximal groove capacity. We propose that the spacer serves to prevent binding of ligands with long lipid tails, whereas short-chain lipids might still displace the PC, which is exposed at the groove entrance. The data presented herein explain how the CD1b groove is preserved, and provide a rationale for the in vivo antigen-binding properties of CD1b.
Figures

References
-
- Batuwangala T, Shepherd D, Gadola SD, Gibson KJ, Zaccai NR, Fersht AR, Besra GS, Cerundolo V, Jones EY (2004) The crystal structure of human CD1b with a bound bacterial glycolipid. J Immunol 172: 2382–2388 - PubMed
-
- Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB (1994) Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature 372: 691–694 - PubMed
-
- Benesch JL, Robinson CV (2006) Mass spectrometry of macromolecular assemblies: preservation and dissociation. Curr Opin Struct Biol 16: 245–251 - PubMed
-
- Brigl M, Brenner MB (2004) CD1: antigen presentation and T cell function. Annu Rev Immunol 22: 817–890 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

