Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1
- PMID: 16874307
- PMCID: PMC1538558
- DOI: 10.1038/sj.emboj.7601239
Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1
Abstract
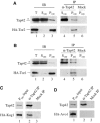
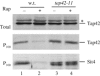
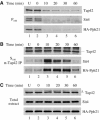
In Saccharomyces cerevisiae, the Tap42-phosphatase complexes are major targets of the Tor kinases in the rapamycin-sensitive signaling pathway. The immunosuppressive agent, rapamycin, induces a prompt activation of the Tap42-associated phosphatases, which is vitally important in Tor-mediated transcriptional regulation. However, the mechanism for the rapid phosphatase activation is poorly understood. In this study, we show that the Tap42-phosphatase complexes exist mainly on membrane structures through their association with Tor complex 1 (TORC1). Rapamycin abrogates this association and releases the Tap42-phosphatase complexes into the cytosol. Disassembly of the Tap42-phosphatase complexes occurs subsequently, following the release but at a much slower rate, presumably caused by Tap42 dephosphorylation. Release of the Tap42-phosphatase complexes from membrane structures also occurs when cells are deprived of nutrient. These findings suggest that the association of the Tap42-phosphatase complexes with TORC1 represents an important mechanism by which nutrient controls Tor signaling activity. In addition, our data support a model in which rapamycin acts not by inhibiting the kinase activity of Tor but by disrupting its interaction with downstream targets.
Figures

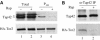




References
-
- Beck T, Hall MN (1999) The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692 - PubMed
-
- Di Como CJ, Arndt KT (1996) Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev 10: 1904–1916 - PubMed
-
- Duvel K, Santhanam A, Garrett S, Schneper L, Broach JR (2003) Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol Cell 11: 1467–1478 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

