The human severe acute respiratory syndrome coronavirus (SARS-CoV) 8b protein is distinct from its counterpart in animal SARS-CoV and down-regulates the expression of the envelope protein in infected cells
- PMID: 16876844
- PMCID: PMC7111915
- DOI: 10.1016/j.virol.2006.06.026
The human severe acute respiratory syndrome coronavirus (SARS-CoV) 8b protein is distinct from its counterpart in animal SARS-CoV and down-regulates the expression of the envelope protein in infected cells
Abstract
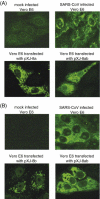
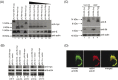
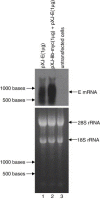
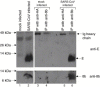
The severe acute respiratory syndrome coronavirus (SARS-CoV), isolated from humans infected during the peak of epidemic, encodes two accessory proteins termed as 8a and 8b. Interestingly, the SARS-CoV isolated from animals contains an extra 29-nucleotide in this region such that these proteins are fused to become a single protein, 8ab. Here, we compared the cellular properties of the 8a, 8b and 8ab proteins by examining their cellular localizations and their abilities to interact with other SARS-CoV proteins. These results may suggest that the conformations of 8a and 8b are different from 8ab although nearly all the amino acids in 8a and 8b are found in 8ab. In addition, the expression of the structural protein, envelope (E), was down-regulated by 8b but not 8a or 8ab. Consequently, E was not detectable in SARS-CoV-infected cells that were expressing high levels of 8b. These findings suggest that 8b may modulate viral replication and/or pathogenesis.
Figures


Similar articles
-
Expression, post-translational modification and biochemical characterization of proteins encoded by subgenomic mRNA8 of the severe acute respiratory syndrome coronavirus.FEBS J. 2007 Aug;274(16):4211-22. doi: 10.1111/j.1742-4658.2007.05947.x. Epub 2007 Jul 20. FEBS J. 2007. PMID: 17645546 Free PMC article.
-
The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8.J Virol. 2007 Dec;81(24):13876-88. doi: 10.1128/JVI.01631-07. Epub 2007 Oct 10. J Virol. 2007. PMID: 17928347 Free PMC article.
-
Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3.Virology. 2018 Feb;515:165-175. doi: 10.1016/j.virol.2017.12.028. Epub 2017 Dec 30. Virology. 2018. PMID: 29294448 Free PMC article.
-
Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus.Antiviral Res. 2006 Nov;72(2):78-88. doi: 10.1016/j.antiviral.2006.05.010. Epub 2006 Jun 6. Antiviral Res. 2006. PMID: 16820226 Free PMC article. Review.
-
SARS coronavirus accessory proteins.Virus Res. 2008 Apr;133(1):113-21. doi: 10.1016/j.virusres.2007.10.009. Epub 2007 Nov 28. Virus Res. 2008. PMID: 18045721 Free PMC article. Review.
Cited by
-
SARS-coronavirus protein 6 conformations required to impede protein import into the nucleus.Virus Res. 2010 Nov;153(2):299-304. doi: 10.1016/j.virusres.2010.08.017. Epub 2010 Aug 26. Virus Res. 2010. PMID: 20800627 Free PMC article.
-
Sequence Analysis of Hot Spot Regions of Spike and RNA-dependent-RNA polymerase (RdRp) Genes of SARS-CoV-2 in Kerman, Iran.Mediterr J Hematol Infect Dis. 2023 Jul 1;15(1):e2023042. doi: 10.4084/MJHID.2023.042. eCollection 2023. Mediterr J Hematol Infect Dis. 2023. PMID: 37435034 Free PMC article.
-
SARS-CoV-2 mutations in Brazil: from genomics to putative clinical conditions.Sci Rep. 2021 Jun 7;11(1):11998. doi: 10.1038/s41598-021-91585-6. Sci Rep. 2021. PMID: 34099808 Free PMC article.
-
Lost in deletion: The enigmatic ORF8 protein of SARS-CoV-2.Biochem Biophys Res Commun. 2021 Jan 29;538:116-124. doi: 10.1016/j.bbrc.2020.10.045. Epub 2020 Oct 21. Biochem Biophys Res Commun. 2021. PMID: 33685621 Free PMC article. Review.
-
Accessory proteins of SARS-CoV and other coronaviruses.Antiviral Res. 2014 Sep;109:97-109. doi: 10.1016/j.antiviral.2014.06.013. Epub 2014 Jul 1. Antiviral Res. 2014. PMID: 24995382 Free PMC article. Review.
References
-
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

