Red blood with blue-blood ancestry: intriguing structure of a snail hemoglobin
- PMID: 16877545
- PMCID: PMC1567689
- DOI: 10.1073/pnas.0601861103
Red blood with blue-blood ancestry: intriguing structure of a snail hemoglobin
Abstract
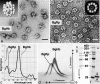
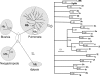
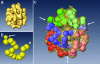
The phylogenetic enigma of snail hemoglobin, its isolated occurrence in a single gastropod family, the Planorbidae, and the lack of sequence data, stimulated the present study. We present here the complete cDNA and predicted amino acid sequence of two hemoglobin polypeptides from the planorbid Biomphalaria glabrata (intermediate host snail for the human parasite Schistosoma mansoni). Both isoforms contain 13 different, cysteine-free globin domains, plus a small N-terminal nonglobin "plug" domain with three cysteines for subunit dimerization (total M(r) approximately 238 kDa). We also identified the native hemoglobin molecule and present here a preliminary 3D reconstruction from electron microscopical images (3 nm resolution); it suggests a 3 x 2-mer quaternary structure (M(r) approximately 1.43 MDa). Moreover, we identified a previously undescribed rosette-like hemolymph protein that has been mistaken for hemoglobin. We also detected expression of an incomplete hemocyanin as trace component. The combined data show that B. glabrata hemoglobin evolved from pulmonate myoglobin, possibly to replace a less-efficient hemocyanin, and reveals a surprisingly simple evolutionary mechanism to create a high molecular mass respiratory protein from 78 similar globin domains.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


Similar articles
-
Recombinant functional multidomain hemoglobin from the gastropod Biomphalaria glabrata.IUBMB Life. 2011 May;63(5):323-8. doi: 10.1002/iub.453. Epub 2011 Apr 13. IUBMB Life. 2011. PMID: 21491558
-
Representation of an immune responsive gene family encoding fibrinogen-related proteins in the freshwater mollusc Biomphalaria glabrata, an intermediate host for Schistosoma mansoni.Gene. 2004 Oct 27;341:255-66. doi: 10.1016/j.gene.2004.07.003. Gene. 2004. PMID: 15474308 Free PMC article.
-
Structural analysis of hemolymph proteins from Schistosoma mansoni (Trematoda)-susceptible and resistant Biomphalaria glabrata (Gastropoda).Comp Biochem Physiol B. 1989;94(3):543-53. doi: 10.1016/0305-0491(89)90194-6. Comp Biochem Physiol B. 1989. PMID: 2620499
-
Molecular studies of Biomphalaria glabrata, an intermediate host of Schistosoma mansoni.Int J Parasitol. 2000 Apr 10;30(4):535-41. doi: 10.1016/s0020-7519(99)00182-4. Int J Parasitol. 2000. PMID: 10731574 Review.
-
Schistosoma mansoni and Biomphalaria: past history and future trends.Parasitology. 2001;123 Suppl:S211-28. doi: 10.1017/s0031182001007703. Parasitology. 2001. PMID: 11769285 Review.
Cited by
-
ProteoPlex: stability optimization of macromolecular complexes by sparse-matrix screening of chemical space.Nat Methods. 2015 Sep;12(9):859-65. doi: 10.1038/nmeth.3493. Epub 2015 Aug 3. Nat Methods. 2015. PMID: 26237227 Free PMC article.
-
Early differential gene expression in haemocytes from resistant and susceptible Biomphalaria glabrata strains in response to Schistosoma mansoni.PLoS One. 2012;7(12):e51102. doi: 10.1371/journal.pone.0051102. Epub 2012 Dec 26. PLoS One. 2012. PMID: 23300533 Free PMC article.
-
Distribution and characterization of rhogocyte cell types in the mantle tissue of Haliotis laevigata.Mar Biotechnol (NY). 2015 Apr;17(2):168-79. doi: 10.1007/s10126-014-9605-9. Epub 2014 Nov 11. Mar Biotechnol (NY). 2015. PMID: 25382219
-
On the Ultrastructure and Function of Rhogocytes from the Pond Snail Lymnaea stagnalis.PLoS One. 2015 Oct 21;10(10):e0141195. doi: 10.1371/journal.pone.0141195. eCollection 2015. PLoS One. 2015. PMID: 26488403 Free PMC article.
-
CryoEM structure and Alphafold molecular modelling of a novel molluscan hemocyanin.PLoS One. 2023 Jun 22;18(6):e0287294. doi: 10.1371/journal.pone.0287294. eCollection 2023. PLoS One. 2023. PMID: 37347755 Free PMC article.
References
-
- Lieb B., Altenhein B., Markl J. J. Biol. Chem. 2000;275:5675–5681. - PubMed
-
- Meissner U., Dube P., Harris J. R., Stark H., Markl J. J. Mol. Biol. 2000;298:21–34. - PubMed
-
- Lieb B., Boisguérin V., Gebauer W., Markl J. J. Mol. Evol. 2004;59:536–545. - PubMed
-
- Figueiredo E. A., Gomes M. V., Heneine I. F., Santos I. O., Hargreaves F. B. Comp. Biochem. Physiol. B. 1973;44:481–491.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

