Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster
- PMID: 16878133
- PMCID: PMC2669546
- DOI: 10.1038/ng1855
Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster
Abstract
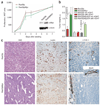
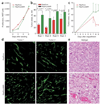
Human adenocarcinomas commonly harbor mutations in the KRAS and MYC proto-oncogenes and the TP53 tumor suppressor gene. All three genetic lesions are potentially pro-angiogenic, as they sustain production of vascular endothelial growth factor (VEGF). Yet Kras-transformed mouse colonocytes lacking p53 formed indolent, poorly vascularized tumors, whereas additional transduction with a Myc-encoding retrovirus promoted vigorous vascularization and growth. In addition, VEGF levels were unaffected by Myc, but enhanced neovascularization correlated with downregulation of anti-angiogenic thrombospondin-1 (Tsp1) and related proteins, such as connective tissue growth factor (CTGF). Both Tsp1 and CTGF are predicted targets for repression by the miR-17-92 microRNA cluster, which was upregulated in colonocytes coexpressing K-Ras and c-Myc. Indeed, miR-17-92 knockdown with antisense 2'-O-methyl oligoribonucleotides partly restored Tsp1 and CTGF expression; in addition, transduction of Ras-only cells with a miR-17-92-encoding retrovirus reduced Tsp1 and CTGF levels. Notably, miR-17-92-transduced cells formed larger, better-perfused tumors. These findings establish a role for microRNAs in non-cell-autonomous Myc-induced tumor phenotypes.
Figures


References
-
- Pelengaris S, Littlewood T, Khan M, Elia G, Evan GI. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol. Cell. 1999;3:565–577. - PubMed
-
- Brandvold KA, Neiman P, Ruddell A. Angiogenesis is an early event in the generation of myc-induced lymphomas. Oncogene. 2000;19:2780–2785. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

