A randomised trial comparing genotypic and virtual phenotypic interpretation of HIV drug resistance: the CREST study
- PMID: 16878178
- PMCID: PMC1523224
- DOI: 10.1371/journal.pctr.0010018
A randomised trial comparing genotypic and virtual phenotypic interpretation of HIV drug resistance: the CREST study
Abstract
Objectives: The aim of this study was to compare the efficacy of different HIV drug resistance test reports (genotype and virtual phenotype) in patients who were changing their antiretroviral therapy (ART).
Design: Randomised, open-label trial with 48-week followup.
Setting: The study was conducted in a network of primary healthcare sites in Australia and New Zealand.
Participants: Patients failing current ART with plasma HIV RNA > 2000 copies/mL who wished to change their current ART were eligible. Subjects were required to be > 18 years of age, previously treated with ART, have no intercurrent illnesses requiring active therapy, and to have provided written informed consent.
Interventions: Eligible subjects were randomly assigned to receive a genotype (group A) or genotype plus virtual phenotype (group B) prior to selection of their new antiretroviral regimen.
Outcome measures: Patient groups were compared for patterns of ART selection and surrogate outcomes (plasma viral load and CD4 counts) on an intention-to-treat basis over a 48-week period.
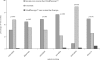
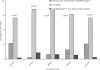
Results: Three hundred and twenty seven patients completing >or= one month of followup were included in these analyses. Resistance tests were the primary means by which ART regimens were selected (group A: 64%, group B: 62%; p = 0.32). At 48 weeks, there were no significant differences between the groups for mean change from baseline plasma HIV RNA (group A: 0.68 log copies/mL, group B: 0.58 log copies/mL; p = 0.23) and mean change from baseline CD4+ cell count (group A: 37 cells/mm(3), group B: 50 cells/mm(3); p = 0.28).
Conclusions: In the absence of clear demonstrated benefits arising from the use of the virtual phenotype interpretation, this study suggests resistance testing using genotyping linked to a reliable interpretive algorithm is adequate for the management of HIV infection.
Figures
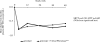

References
-
- Larder BA, Darby G, Richman D. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243:1731–1734. - PubMed
-
- Smith M, Salomon H, Wainberg MA. Development and significance of nucleoside drug resistance in infection caused by the human immunodeficiency virus type 1. Clin Invest Med. 1994;17((3)):226–243. - PubMed
-
- Mayers DL. Prevalence and incidence of resistance to zidovudine and other antiretroviral drugs. Am J Med. 1997;102:70–75. - PubMed
-
- Condra JH, Schleif WA, Blahy OM, et al. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. - PubMed
-
- Hirsch MS, Conway B, D'Aquila RT, et al. Antiretroviral drug resistance testing in adults with HIV infection: Implications for clinical management. International AIDS Society—USA Panel. JAMA. 1998;24:1984–1991. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

