Breakdown of the reciprocal stabilization of QBRICK/Frem1, Fras1, and Frem2 at the basement membrane provokes Fraser syndrome-like defects
- PMID: 16880404
- PMCID: PMC1567684
- DOI: 10.1073/pnas.0601011103
Breakdown of the reciprocal stabilization of QBRICK/Frem1, Fras1, and Frem2 at the basement membrane provokes Fraser syndrome-like defects
Abstract
An emerging family of extracellular matrix proteins characterized by 12 consecutive CSPG repeats and the presence of Calx-beta motif(s) includes Fras1, QBRICK/Frem1, and Frem2. Mutations in the genes encoding these proteins have been associated with mouse models of Fraser syndrome, which is characterized by subepidermal blistering, cryptophthalmos, syndactyly, and renal dysmorphogenesis. Here, we report that all of these proteins are localized to the basement membrane, and that their basement membrane localization is simultaneously impaired in Fraser syndrome model mice. In Frem2 mutant mice, not only Frem2 but Fras1 and QBRICK/Frem1 were depleted from the basement membrane zone. This coordinated reduction in basement membrane deposition was also observed in another Fraser syndrome model mouse, in which GRIP1, a Fras1- and Frem2-interacting adaptor protein, is primarily affected. Targeted disruption of Qbrick/Frem1 also resulted in diminished expression of Fras1 and Frem2 at the epidermal basement membrane, confirming the reciprocal stabilization of QBRICK/Frem1, Fras1, and Frem2 in this location. When expressed and secreted by transfected cells, these proteins formed a ternary complex, raising the possibility that their reciprocal stabilization at the basement membrane is due to complex formation. Given the close association of Fraser syndrome phenotypes with defective epidermal-dermal interactions, the coordinated assembly of three Fraser syndrome-associated proteins at the basement membrane appears to be instrumental in epidermal-dermal interactions during morphogenetic processes.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


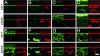


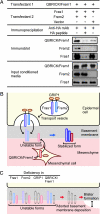
Similar articles
-
Frem3, a member of the 12 CSPG repeats-containing extracellular matrix protein family, is a basement membrane protein with tissue distribution patterns distinct from those of Fras1, Frem2, and QBRICK/Frem1.Matrix Biol. 2007 Jul;26(6):456-62. doi: 10.1016/j.matbio.2007.03.001. Epub 2007 Mar 30. Matrix Biol. 2007. PMID: 17462874
-
Basement membrane localization of Frem3 is independent of the Fras1/Frem1/Frem2 protein complex within the sublamina densa.Matrix Biol. 2007 Oct;26(8):652-8. doi: 10.1016/j.matbio.2007.05.008. Epub 2007 Jun 6. Matrix Biol. 2007. PMID: 17596926
-
The role of Fras1/Frem proteins in the structure and function of basement membrane.Int J Biochem Cell Biol. 2011 Apr;43(4):487-95. doi: 10.1016/j.biocel.2010.12.016. Epub 2010 Dec 21. Int J Biochem Cell Biol. 2011. PMID: 21182980 Review.
-
The Fras1/Frem family of extracellular matrix proteins: structure, function, and association with Fraser syndrome and the mouse bleb phenotype.Connect Tissue Res. 2008;49(3):277-82. doi: 10.1080/03008200802148025. Connect Tissue Res. 2008. PMID: 18661360
-
Let's stick together: the role of the Fras1 and Frem proteins in epidermal adhesion.IUBMB Life. 2007 Jul;59(7):427-35. doi: 10.1080/15216540701510581. IUBMB Life. 2007. PMID: 17654118 Review.
Cited by
-
Novel frem1-related mouse phenotypes and evidence of genetic interactions with gata4 and slit3.PLoS One. 2013;8(3):e58830. doi: 10.1371/journal.pone.0058830. Epub 2013 Mar 11. PLoS One. 2013. PMID: 23536828 Free PMC article.
-
Identification and Comprehensive Analysis of FREM2 Mutation as a Potential Prognostic Biomarker in Colorectal Cancer.Front Mol Biosci. 2022 Feb 18;9:839617. doi: 10.3389/fmolb.2022.839617. eCollection 2022. Front Mol Biosci. 2022. PMID: 35252356 Free PMC article.
-
Fraser syndrome in three consecutive siblings.Oman J Ophthalmol. 2011 May;4(2):87-9. doi: 10.4103/0974-620X.83661. Oman J Ophthalmol. 2011. PMID: 21897626 Free PMC article.
-
A homozygous mutation p.Arg2167Trp in FREM2 causes isolated cryptophthalmos.Hum Mol Genet. 2018 Jul 1;27(13):2357-2366. doi: 10.1093/hmg/ddy144. Hum Mol Genet. 2018. PMID: 29688405 Free PMC article.
-
Disease mechanisms of monogenic congenital anomalies of the kidney and urinary tract American Journal of Medical Genetics Part C.Am J Med Genet C Semin Med Genet. 2022 Sep;190(3):325-343. doi: 10.1002/ajmg.c.32006. Epub 2022 Oct 8. Am J Med Genet C Semin Med Genet. 2022. PMID: 36208064 Free PMC article. Review.
References
-
- Boyd P. A., Keeling J. W., Lindenbaum R. H. Am. J. Med. Genet. 1988;31:159–168. - PubMed
-
- Darling S., Gossler A. Clin. Dysmorphol. 1994;3:91–95. - PubMed
-
- McGregor L., Makela V., Darling S. M., Vrontou S., Chalepakis G., Roberts C., Smart N., Rutland P., Prescott N., Hopkins J., et al. Nat. Genet. 2003;34:203–208. - PubMed
-
- Jadeja S., Smyth I., Pitera J. E., Taylor M. S., van Haelst M., Bentley E., McGregor L., Hopkins J., Chalepakis G., Philip N., et al. Nat. Genet. 2005;37:520–525. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

