Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene
- PMID: 16880520
- PMCID: PMC1592793
- DOI: 10.1128/MCB.02366-05
Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene
Abstract
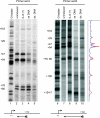
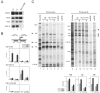
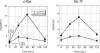
Human 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole sensitivity-inducing factor (DSIF) and negative elongation factor (NELF) negatively regulate transcription elongation by RNA polymerase II (RNAPII) in vitro. However, the physiological roles of this negative regulation are not well understood. Here, by using a number of approaches to identify protein-DNA interactions in vivo, we show that DSIF- and NELF-mediated transcriptional pausing has a dual function in regulating immediate-early expression of the human junB gene. Before induction by interleukin-6, RNAPII, DSIF, and NELF accumulate in the promoter-proximal region of junB, mainly at around position +50 from the transcription initiation site. After induction, the association of these proteins with the promoter-proximal region continues whereas RNAPII and DSIF are also found in the downstream regions. Depletion of a subunit of NELF by RNA interference enhances the junB mRNA level both before and after induction, indicating that DSIF- and NELF-mediated pausing contributes to the negative regulation of junB expression, not only by inducing RNAPII pausing before induction but also by attenuating transcription after induction. These regulatory mechanisms appear to be conserved in other immediate-early genes as well.
Figures




References
-
- Abe, K., M. Hirai, K. Mizuno, N. Higashi, T. Sekimoto, T. Miki, T. Hirano, and K. Nakajima. 2001. The YXXQ motif in gp130 is crucial for STAT3 phosphorylation at Ser727 through an H7-sensitive kinase pathway. Oncogene 20:3464-3474. - PubMed
-
- Adelman, K., M. T. Marr, J. Werner, A. Saunders, Z. Ni, E. D. Andrulis, and J. T. Lis. 2005. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell 17:103-112. - PubMed
-
- Aiyar, S. E., J. L. Sun, A. L. Blair, C. A. Moskaluk, Y. Z. Lu, Q. N. Ye, Y. Yamaguchi, A. Mukherjee, D. M. Ren, H. Handa, and R. Li. 2004. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 18:2134-2146. - PMC - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
