Nef interference with HIV-1-specific CTL antiviral activity is epitope specific
- PMID: 16882705
- PMCID: PMC1895430
- DOI: 10.1182/blood-2006-06-030668
Nef interference with HIV-1-specific CTL antiviral activity is epitope specific
Abstract
HIV-1 Nef and HIV-1-specific cytotoxic T lymphocytes (CTLs) have important and opposing roles in the immunopathogenesis of HIV-1 infection. Nef-mediated down-modulation of HLA class I on infected cells can confer resistance to CTL clearance, but the factors determining the efficiency of this process are unknown. This study examines the impact of Nef on the antiviral activity of several CTL clones recognizing epitopes from early and late HIV-1 proteins, restricted by HLA-A, -B, and -C molecules. CTL-targeting epitopes in early proteins remained susceptible to the effects of Nef, although possibly to a lesser degree than CTL-targeting late protein epitopes, indicating that significant Nef-mediated HLA down-regulation can precede even the presentation of early protein-derived epitopes. However, HLA-C-restricted CTLs were unaffected by Nef, consistent with down-regulation of cell-surface HLA-A and -B but not HLA-C molecules. Thus, CTLs vary dramatically in their susceptibility to Nef interference, suggesting differences in the relative importance of HLA-A- and HLA-B- versus HLA-C-restricted CTLs in vivo. The data thus indicate that HLA-C-restricted CTLs may have an under-appreciated antiviral role in the setting of Nef in vivo and suggest a benefit of promoting HLA-C-restricted CTLs for immunotherapy or vaccine development.
Figures


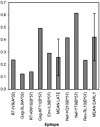

References
-
- Piguet V, Trono D. A structure-function analysis of the nef protein of primate lentiviruses. In: Kuiken C, Foley B, Hahn B, et al, eds. Human Retroviruses and AIDS 1999. Los Alamos, NM: Los Alamos National Laboratory; 1999: 448-459.
-
- Oelrichs R, Tsykin A, Rhodes D, et al. Genomic sequence of HIV type 1 from four members of the Sydney Blood Bank Cohort of long-term nonprogressors. AIDS Res Hum Retroviruses. 1998;14: 811-814. - PubMed
-
- Daniel MD, Kirchhoff F, Czajak SC, Sehgal PK, Desrosiers RC. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258: 1938-1941. - PubMed
-
- Yang OO, Walker BD. CD8+ cells in human immunodeficiency virus type I pathogenesis: cytolytic and noncytolytic inhibition of viral replication. Adv Immunol. 1997;66: 273-311. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

