Reconstitution of paired T cell receptor alpha- and beta-chains from microdissected single cells of human inflammatory tissues
- PMID: 16882720
- PMCID: PMC1567696
- DOI: 10.1073/pnas.0604247103
Reconstitution of paired T cell receptor alpha- and beta-chains from microdissected single cells of human inflammatory tissues
Abstract
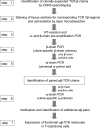
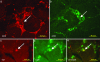
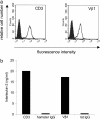
We describe a strategy to "revive" putatively pathogenic T cells from frozen specimens of human inflammatory target organs. To distinguish pathogenic from irrelevant bystander T cells, we focused on cells that were (i) clonally expanded and (ii) in direct morphological contact with a target cell. Using CDR3 spectratyping, we identified clonally expanded T cell receptor (TCR) beta-chains in muscle sections of patients with inflammatory muscle diseases. By immunohistochemistry, we identified those Vbeta-positive T cells that fulfilled the morphological criteria of myocytotoxicity and isolated them by laser microdissection. Next, we identified coexpressed pairs of TCR alpha- and beta-chains by a multiplex PCR protocol, which allows the concomitant amplification of both chains from single cells. This concomitant amplification had not been achieved previously in histological sections, mainly because of the paucity of available anti-alpha-chain antibodies and the great heterogeneity of the alpha-chain genes. From muscle tissue of a patient with polymyositis, we isolated 64 T cells that expressed an expanded Vbeta1 chain. In 23 of these cells, we identified the corresponding alpha-chain. Twenty of these 23 alpha-chains were identical, suggesting antigen-driven selection. After functional reconstitution of the alphabeta-pairs, their antigen-recognition properties could be studied. Our results open avenues for combined analysis of the full TCR alpha- and beta-chain repertoire in human inflammatory tissues.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

