The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components
- PMID: 16882981
- PMCID: PMC1536055
- DOI: 10.1101/gad.386106
The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components
Abstract
Regulation of ribosome biogenesis is a key element of cell biology, not only because ribosomes are directly required for growth, but also because ribosome production monopolizes nearly 80% of the global transcriptional activity in rapidly growing yeast cells. These observations underscore the need for a tight regulation of ribosome synthesis in response to environmental conditions. In eukaryotic cells, ribosome synthesis involves the activities of the three nuclear RNA polymerases (Pol). Although postulated, there is no clear evidence indicating whether the maintenance of an equimolar supply of ribosomal components reflects communication between the nuclear transcriptional machineries. Here, by constructing a yeast strain expressing a Pol I that remains constitutively competent for the initiation of transcription under stress conditions, we demonstrate that derepression of Pol I transcription leads to a derepression of Pol II transcription that is restricted to the genes encoding ribosomal proteins. Furthermore, we show that the level of 5S rRNA, synthesized by Pol III, is deregulated concomitantly with Pol I transcription. Altogether, these results indicate that a partial derepression of Pol I activity drives an abnormal accumulation of all ribosomal components, highlighting the critical role of the regulation of Pol I activity within the control of ribosome biogenesis.
Figures

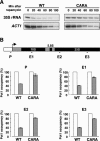

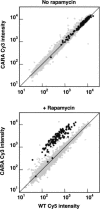
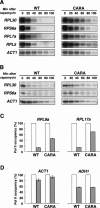


Comment in
-
Does Pol I talk to Pol II? Coordination of RNA polymerases in ribosome biogenesis.Genes Dev. 2006 Aug 1;20(15):1982-5. doi: 10.1101/gad.1460706. Genes Dev. 2006. PMID: 16882974 No abstract available.
References
-
- Clarke E.M., Peterson C.L., Brainard A.V., Riggs D.L. Regulation of the RNA polymerase I and III transcription systems in response to growth conditions. J. Biol. Chem. 1996;27:22189–22195. - PubMed
-
- Claypool J.A., French S.L., Johzuka K., Eliason K., Vu L., Dodd J.A., Beyer A.L., Nomura M. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol. Biol. Cell. 2004;15:946–956. - PMC - PubMed
-
- Cormack B.P., Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
