E4F1: a novel candidate factor for mediating BMI1 function in primitive hematopoietic cells
- PMID: 16882984
- PMCID: PMC1536061
- DOI: 10.1101/gad.1453406
E4F1: a novel candidate factor for mediating BMI1 function in primitive hematopoietic cells
Abstract
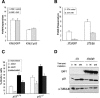
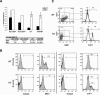
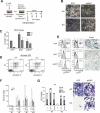
The Polycomb group gene Bmi1 is essential for the proliferation of neural and hematopoietic stem cells. Much remains to be learned about the pathways involved in the severe hematopoietic phenotype observed in Bmi1 homozygous mutant mice except for the fact that loss of p53 or concomitant loss of p16(Ink4a) and p19(Arf) functions achieves only a partial rescue. Here we report the identification of E4F1, an inhibitor of cellular proliferation, as a novel BMI1-interacting partner in hematopoietic cells. We provide evidence that Bmi1 and E4f1 genetically interact in the hematopoietic compartment to regulate cellular proliferation. Most importantly, we demonstrate that reduction of E4f1 levels through RNA interference mediated knockdown is sufficient to rescue the clonogenic and repopulating ability of Bmi1(-/-) hematopoietic cells up to 3 mo post-transplantation. Using cell lines and MEF, we also demonstrate that INK4A/ARF and p53 are not essential for functional interaction between Bmi1 and E4f1. Together, these findings identify E4F1 as a key modulator of BMI1 activity in primitive hematopoietic cells.
Figures



References
-
- Ahmed-Choudhury J., Agathanggelou A., Fenton S.L., Ricketts C., Clark G.J., Maher E.R., Latif F. Transcriptional regulation of cyclin A2 by RASSF1A through the enhanced binding of p120E4F to the cyclin A2 promoter. Cancer Res. 2005;65:2690–2697. - PubMed
-
- Benveniste P., Cantin C., Hyam D., Iscove N.N. Hematopoietic stem cells engraft in mice with absolute efficiency. Nat. Immunol. 2003;4:708–713. - PubMed
-
- Bruggeman S.W., Valk-Lingbeek M.E., van der Stoop P.P., Jacobs J.J., Kieboom K., Tanger E., Hulsman D., Leung C., Arsenijevic Y., Marino S., et al. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes & Dev. 2005;19:1438–1443. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
