Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes
- PMID: 16884331
- PMCID: PMC1526461
- DOI: 10.1371/journal.pcbi.0020100
Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes
Abstract
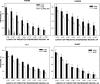
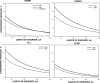
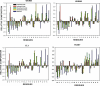
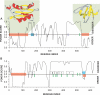
Recent proteome-wide screening approaches have provided a wealth of information about interacting proteins in various organisms. To test for a potential association between protein connectivity and the amount of predicted structural disorder, the disorder propensities of proteins with various numbers of interacting partners from four eukaryotic organisms (Caenorhabditis elegans, Saccharomyces cerevisiae, Drosophila melanogaster, and Homo sapiens) were investigated. The results of PONDR VL-XT disorder analysis show that for all four studied organisms, hub proteins, defined here as those that interact with > or = 10 partners, are significantly more disordered than end proteins, defined here as those that interact with just one partner. The proportion of predicted disordered residues, the average disorder score, and the number of predicted disordered regions of various lengths were higher overall in hubs than in ends. A binary classification of hubs and ends into ordered and disordered subclasses using the consensus prediction method showed a significant enrichment of wholly disordered proteins and a significant depletion of wholly ordered proteins in hubs relative to ends in worm, fly, and human. The functional annotation of yeast hubs and ends using GO categories and the correlation of these annotations with disorder predictions demonstrate that proteins with regulation, transcription, and development annotations are enriched in disorder, whereas proteins with catalytic activity, transport, and membrane localization annotations are depleted in disorder. The results of this study demonstrate that intrinsic structural disorder is a distinctive and common characteristic of eukaryotic hub proteins, and that disorder may serve as a determinant of protein interactivity.
Conflict of interest statement
Figures

References
-
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae . Nature. 2000;403:623–627. - PubMed
-
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, et al. A protein interaction map of Drosophila melanogaster . Science. 2003;302:1727–1736. - PubMed
-
- Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, et al. The protein–protein interaction map of Helicobacter pylori . Nature. 2001;409:211–215. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

