Activation of heterotrimeric G proteins by Smoothened
- PMID: 16885213
- PMCID: PMC1567926
- DOI: 10.1073/pnas.0600880103
Activation of heterotrimeric G proteins by Smoothened
Abstract
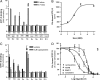
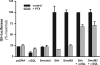
The mechanisms by which the activation of Smoothened (Smo), a protein essential to the actions of the Hedgehog family of secreted proteins, is translated into signals that converge on the Gli transcription factors are not fully understood. The seven-transmembrane structure of Smo has long implied the utilization of heterotrimeric GTP-binding regulatory proteins (G proteins); however, evidence in this regard has been indirect and contradictory. In the current study we evaluated the capacity of mammalian Smo to couple to G proteins directly. We found that Smo, by virtue of what appears to be constitutive activity, activates all members of the G(i) family but does not activate members of the G(s), G(q), and G(12) families. The activation is suppressed by cyclopamine and other inhibitors of Hedgehog signaling and is enhanced by the Smo agonist purmorphamine. Activation of G(i) by Smo is essential in the activation of Gli in fibroblasts, because disruption of coupling to G(i) with pertussis toxin inhibits the activation of Gli by Sonic hedgehog and a constitutively active form of Smo (SmoM2). However, G(i) does not provide a sufficient signal because a truncated form of Smo, although capable of activating G(i), does not effect activation of Gli. Rescue of pertussis toxin-inhibited activation of Gli by Sonic hedgehog can be achieved with a constitutively active Galpha(i)-subunit. The data suggest that Smo is in fact the source of two signals relevant to the activation of Gli: one involving G(i) and the other involving events at Smo's C-tail independent of G(i).
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


References
-
- Ingham P. W., McMahon A. P. Genes Dev. 2001;15:3059–3087. - PubMed
-
- Nieuwenhuis E., Hui C.-c. Clin. Genet. 2005;67:193–208. - PubMed
-
- di Magliano M. P., Hebrok M. Nat. Rev. Cancer. 2003;3:903–911. - PubMed
-
- Stone D. M., Hynes M., Armanini M., Swanson T. A., Gu Q., Johnson R. L., Scott M. P., Pennica D., Goddard A., Phillips H., et al. Nature. 1996;384:129–134. - PubMed
-
- Wang B., Fallon J. F., Beachy P. A. Cell. 2000;100:423–434. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

