Peptide aMptD-mediated capture PCR for detection of Mycobacterium avium subsp. paratuberculosis in bulk milk samples
- PMID: 16885259
- PMCID: PMC1538760
- DOI: 10.1128/AEM.00590-06
Peptide aMptD-mediated capture PCR for detection of Mycobacterium avium subsp. paratuberculosis in bulk milk samples
Abstract
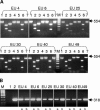
A peptide-mediated capture PCR for the detection of Mycobacterium avium subsp. paratuberculosis in bulk milk samples was developed and characterized. Capture of the organism was performed using peptide aMptD, which had been shown to bind to the M. avium subsp. paratuberculosis MptD protein (J. Stratmann, B. Strommenger, R. Goethe, K. Dohmann, G. F. Gerlach, K. Stevenson, L. L. Li, Q. Zhang, V. Kapur, and T. J. Bull, Infect. Immun. 72:1265-1274, 2004). Consistent expression of the MptD receptor protein and binding of the aMptD ligand were demonstrated by capturing different Mycobacterium avium subsp. paratuberculosis type I and type II strains and subsequent PCR analysis using ISMav2-based primers. The analytical sensitivity of the method was determined to be 5 x 10(2) CFU ml(-1) for artificially contaminated milk. The specificity of aMptD binding was confirmed by culture and competitive capture assays, showing selective enrichment of M. avium subsp. paratuberculosis (at a concentration of 5 x 10(2) CFU ml(-1)) from samples containing 100- and 1,000-fold excesses of other mycobacterial species, including M. avium subsp. avium and M. avium subsp. hominissuis. The aMptD-mediated capture of M. avium subsp. paratuberculosis using paramagnetic beads, followed by culture, demonstrated the ability of this approach to capture viable target cells present in artificially contaminated milk. Surface plasmon resonance experiments revealed that the aMptD peptide is a high-affinity ligand with a calculated association rate constant of 9.28 x 10(3) and an association constant of 1.33 x 10(9). The potential use of the method on untreated raw milk in the field was investigated by testing 423 bulk milk samples obtained from different dairy farms in Germany, 23 of which tested positive. Taken together, the results imply that the peptide-mediated capture PCR might present a suitable test for paratuberculosis screening of dairy herds, as it has an analytical sensitivity sufficient for detection of M. avium subsp. paratuberculosis in bulk milk samples under field conditions, relies on a defined and validated ligand-receptor interaction, and is adaptable to routine diagnostic laboratory automation.
Figures

Similar articles
-
Automated high-throughput immunomagnetic separation-PCR for detection of Mycobacterium avium subsp. paratuberculosis in bovine milk.Int J Food Microbiol. 2006 Aug 1;110(3):201-8. doi: 10.1016/j.ijfoodmicro.2006.01.042. Epub 2006 Jul 11. Int J Food Microbiol. 2006. PMID: 16814891
-
Development of a peptide-mediated capture PCR for detection of Mycobacterium avium subsp. paratuberculosis in milk.J Clin Microbiol. 2002 Nov;40(11):4244-50. doi: 10.1128/JCM.40.11.4244-4250.2002. J Clin Microbiol. 2002. PMID: 12409405 Free PMC article.
-
Mycobacterium avium subsp. paratuberculosis detection in individual and bulk tank milk samples from bovine herds and caprine flocks.Foodborne Pathog Dis. 2010 Apr;7(4):351-5. doi: 10.1089/fpd.2009.0374. Foodborne Pathog Dis. 2010. PMID: 19911881
-
Magnetic Separation Methods for the Detection of Mycobacterium avium subsp. paratuberculosis in Various Types of Matrices: A Review.Biomed Res Int. 2017;2017:5869854. doi: 10.1155/2017/5869854. Epub 2017 May 31. Biomed Res Int. 2017. PMID: 28642876 Free PMC article. Review.
-
A scoping review of the testing of bulk milk to detect infectious diseases of dairy cattle: Diseases caused by bacteria.J Dairy Sci. 2023 Mar;106(3):1986-2006. doi: 10.3168/jds.2022-22395. Epub 2023 Jan 27. J Dairy Sci. 2023. PMID: 36710183
Cited by
-
Enhanced expression of codon optimized Mycobacterium avium subsp. paratuberculosis antigens in Lactobacillus salivarius.Front Cell Infect Microbiol. 2014 Sep 4;4:120. doi: 10.3389/fcimb.2014.00120. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25237653 Free PMC article.
-
Mycobacterium paratuberculosis detection in cow's milk in Argentina by immunomagnetic separation-PCR.Braz J Microbiol. 2016 Apr-Jun;47(2):506-12. doi: 10.1016/j.bjm.2016.01.013. Epub 2016 Mar 2. Braz J Microbiol. 2016. PMID: 26991290 Free PMC article.
-
Is the transmission of Mycobacterium avium subspecies paratuberculosis (MAP) infection through milk intended to feed calves an overlooked item in paratuberculosis control programs?Trop Anim Health Prod. 2020 Jan;52(1):89-94. doi: 10.1007/s11250-019-01988-x. Epub 2019 Jul 19. Trop Anim Health Prod. 2020. PMID: 31325018
-
Production and evaluation of antibodies and phage display-derived peptide ligands for immunomagnetic separation of Mycobacterium bovis.J Clin Microbiol. 2012 May;50(5):1598-605. doi: 10.1128/JCM.05747-11. Epub 2012 Feb 8. J Clin Microbiol. 2012. PMID: 22322353 Free PMC article.
-
Maximizing capture efficiency and specificity of magnetic separation for Mycobacterium avium subsp. paratuberculosis cells.Appl Environ Microbiol. 2010 Nov;76(22):7550-8. doi: 10.1128/AEM.01432-10. Epub 2010 Sep 17. Appl Environ Microbiol. 2010. PMID: 20851966 Free PMC article.
References
-
- Benedictus, G., and C. J. Kalis. 2003. Paratuberculosis: eradication, control and diagnostic methods. Acta Vet. Scand. 44:231-241. - PubMed
-
- Beyerbach, M., T. Rehm, L. Kreienbrock, and G. F. Gerlach. 2001. Eradication of paratuberculosis in dairy herds: determination of the initial herd prevalence and modelling of prevalence development. Dtsch. Tierarztl. Wochenschr. 108:291-296. - PubMed
-
- Bitsch, V., and L. Ronsholt. 1995. Control of bovine viral diarrhea virus infection without vaccines. Vet. Clin. N. Am. Food Anim. Pract. 11:627-640. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

