SigmaB activation under environmental and energy stress conditions in Listeria monocytogenes
- PMID: 16885265
- PMCID: PMC1538764
- DOI: 10.1128/AEM.03058-05
SigmaB activation under environmental and energy stress conditions in Listeria monocytogenes
Abstract
To measure sigmaB activation in Listeria monocytogenes under environmental or energy stress conditions, quantitative reverse transcriptase PCR (TaqMan) was used to determine the levels of transcripts for the sigmaB -dependent opuCA and clpC genes in strains having null mutations in genes encoding regulator of sigma B proteins (rsbT and rsbV) and sigma B (sigB) and in the L. monocytogenes wild-type 10403S strain under different stress conditions. The DeltasigB, DeltarsbT, and DeltarsbV strains previously exhibited increased hemolytic activities compared to the hemolytic activity of the wild-type strain; therefore, transcript levels for hly were also determined. RsbT, RsbV, and sigmaB were all required for opuCA expression during growth under carbon-limiting conditions or following exposure to pH 4.5, salt, ethanol, or the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP). Expression of clpC was RsbT, RsbV, and sigmaB dependent in the presence of CCCP but not under the other conditions. hly expression was not RsbT, RsbV, or sigmaB dependent in the presence of either CCCP or salt. opuCA transcript levels did not increase in the presence of rapidly lethal stresses (i.e., pH 2.5 or 13 mM cumene hydroperoxide) despite the enhanced survival of the wild type compared with the survival of the mutant strains under these conditions. These findings highlight the importance of complementing phenotypic characterizations with gene expression studies to identify direct and indirect effects of null mutations in regulatory genes, such as sigB. Overall, our data show that while sigmaB activation occurs through a single pathway under both environmental and energy stress conditions, regulation of expression of some stress response and virulence genes in the sigmaB regulon (e.g., clpC) appears to require networks involving multiple transcriptional regulators.
Figures
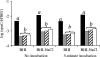

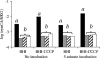
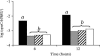

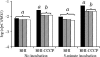
Similar articles
-
RsbT and RsbV contribute to sigmaB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes.Appl Environ Microbiol. 2004 Sep;70(9):5349-56. doi: 10.1128/AEM.70.9.5349-5356.2004. Appl Environ Microbiol. 2004. PMID: 15345420 Free PMC article.
-
Contributions of Listeria monocytogenes sigmaB and PrfA to expression of virulence and stress response genes during extra- and intracellular growth.Microbiology (Reading). 2006 Jun;152(Pt 6):1827-1838. doi: 10.1099/mic.0.28758-0. Microbiology (Reading). 2006. PMID: 16735745
-
SigmaB-dependent and sigmaB-independent mechanisms contribute to transcription of Listeria monocytogenes cold stress genes during cold shock and cold growth.Appl Environ Microbiol. 2007 Oct;73(19):6019-29. doi: 10.1128/AEM.00714-07. Epub 2007 Aug 3. Appl Environ Microbiol. 2007. PMID: 17675428 Free PMC article.
-
Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the sigmaB regulon.Mol Microbiol. 1998 Sep;29(5):1129-36. doi: 10.1046/j.1365-2958.1998.00977.x. Mol Microbiol. 1998. PMID: 9767581 Review.
-
[Contribution of sigma B to environmental stress tolerance in Listeria monocytogenes--a review].Wei Sheng Wu Xue Bao. 2009 Oct;49(10):1282-8. Wei Sheng Wu Xue Bao. 2009. PMID: 20069872 Review. Chinese.
Cited by
-
Characterization of a potential Listeria monocytogenes virulence factor associated with attachment to fresh produce.Appl Environ Microbiol. 2013 Nov;79(22):6855-61. doi: 10.1128/AEM.01006-13. Epub 2013 Aug 23. Appl Environ Microbiol. 2013. PMID: 23974144 Free PMC article.
-
Regulatory network features in Listeria monocytogenes-changing the way we talk.Front Cell Infect Microbiol. 2014 Feb 14;4:14. doi: 10.3389/fcimb.2014.00014. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 24592357 Free PMC article. Review.
-
Blue Light Sensing in Listeria monocytogenes Is Temperature-Dependent and the Transcriptional Response to It Is Predominantly SigB-Dependent.Front Microbiol. 2019 Nov 14;10:2497. doi: 10.3389/fmicb.2019.02497. eCollection 2019. Front Microbiol. 2019. PMID: 31798538 Free PMC article.
-
Differential regulation of Listeria monocytogenes internalin and internalin-like genes by sigmaB and PrfA as revealed by subgenomic microarray analyses.Foodborne Pathog Dis. 2008 Aug;5(4):417-35. doi: 10.1089/fpd.2008.0085. Foodborne Pathog Dis. 2008. PMID: 18713061 Free PMC article.
-
The σB-Mediated General Stress Response of Listeria monocytogenes: Life and Death Decision Making in a Pathogen.Front Microbiol. 2020 Jul 7;11:1505. doi: 10.3389/fmicb.2020.01505. eCollection 2020. Front Microbiol. 2020. PMID: 32733414 Free PMC article. Review.
References
-
- Bernhardt, J., U. Völker, A. Völker, H. Antelmann, R. Schmid, H. Mach, and M. Hecker. 1997. Specific and general stress proteins in. Bacillus subtilis—a two-dimensional protein electrophoresis study. Microbiology 143:999-1017. - PubMed
-
- Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

