DksA is required for growth phase-dependent regulation, growth rate-dependent control, and stringent control of fis expression in Escherichia coli
- PMID: 16885445
- PMCID: PMC1540068
- DOI: 10.1128/JB.00276-06
DksA is required for growth phase-dependent regulation, growth rate-dependent control, and stringent control of fis expression in Escherichia coli
Abstract
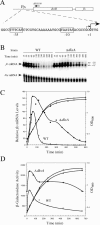
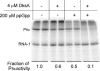
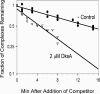
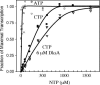
DksA is a critical transcription factor in Escherichia coli that binds to RNA polymerase and potentiates control of rRNA promoters and certain amino acid promoters. Given the kinetic similarities between rRNA promoters and the fis promoter (Pfis), we investigated the possibility that DksA might also control transcription from Pfis. We show that the absence of dksA extends transcription from Pfis well into the late logarithmic and stationary growth phases, demonstrating the importance of DksA for growth phase-dependent regulation of fis. We also show that transcription from Pfis increases with steady-state growth rate and that dksA is absolutely required for this regulation. In addition, both DksA and ppGpp are required for inhibition of Pfis promoter activity following amino acid starvation, and these factors act directly and synergistically to negatively control Pfis transcription in vitro. DksA decreases the half-life of the intrinsically short-lived fis promoter-RNA polymerase complex and increases its sensitivity to the concentration of CTP, the predominant initiating nucleotide triphosphate for this promoter. This work extends our understanding of the multiple factors controlling fis expression and demonstrates the generality of the DksA requirement for regulation of kinetically similar promoters.
Figures


References
-
- Aiyar, S. E., S. M. McLeod, W. Ross, C. A. Hirvonen, M. S. Thomas, R. C. Johnson, and R. L. Gourse. 2002. Architecture of Fis-activated transcription complexes at the Escherichia coli rrnB P1 and rrnE P1 promoters. J. Mol. Biol. 316:501-516. - PubMed
-
- Barker, M. M., T. Gaal, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 305:689-702. - PubMed
-
- Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305:673-688. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

