Characterization of the pilin ortholog of the Helicobacter pylori type IV cag pathogenicity apparatus, a surface-associated protein expressed during infection
- PMID: 16885455
- PMCID: PMC1540075
- DOI: 10.1128/JB.00060-06
Characterization of the pilin ortholog of the Helicobacter pylori type IV cag pathogenicity apparatus, a surface-associated protein expressed during infection
Abstract
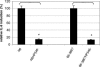
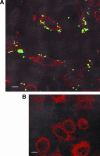
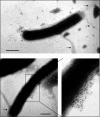
The Helicobacter pylori cag pathogenicity island (cag PAI) encodes components of a type IV secretion system (T4SS) involved in host interaction and pathogenicity. Previously, seven cag PAI proteins were identified as homologs of Agrobacterium tumefaciens Vir proteins, which form a paradigm T4SS. The T pilus composed of the processed VirB2 pilin is an external structural part of the A. tumefaciens T4SS. In H. pylori, cag-dependent assembly of pili has not been observed so far, nor has a pilin (VirB2) ortholog been characterized. We have here identified, using a motif-based search, an H. pylori cag island protein (HP0546) that possesses sequence and predicted structural similarities to VirB2-like pilins of other T4SSs. The HP0546 protein displays interstrain variability in its terminal domains. HP0546 was expressed as a FLAG-tagged fusion protein in Escherichia coli, A. tumefaciens, and H. pylori and was detected as either two or three bands of different molecular masses in the insoluble fraction, indicating protein processing. As reported previously, isogenic H. pylori mutants in the putative cag pilin gene had reduced abilities to induce cag PAI-dependent interleukin-8 secretion in gastric epithelial cells. Fractionation analysis of H. pylori, using a specific antiserum raised against an N-terminal HP0546 peptide, showed that the protein is partially surface exposed and that its surface localization depended upon an intact cag system. By immunoelectron microscopy, HP0546 was localized in surface appendages, with surface exposure of an N-terminal epitope. Pronounced strain-to-strain variability of this predicted surface-exposed part of HP0546 indicates a strong selective pressure for variation in vivo.
Figures



References
-
- Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol. 28:37-53. - PubMed
-
- Alm, R. A., L.-S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. - PubMed
-
- Backert, S., E. Ziska, V. Brinkmann, U. Zimny-Arndt, A. Fauconnier, P. R. Jungblut, M. Naumann, and T. F. Meyer. 2000. Translocation of the Helicobacter pylori CagA protein in gastric epithelial cells by a type IV secretion apparatus. Cell. Microbiol. 2:155-164. - PubMed
-
- Buhrdorf, R., C. Forster, R. Haas, and W. Fischer. 2003. Topological analysis of a putative virB8 homologue essential for the cag type IV secretion system in Helicobacter pylori. Int. J. Med. Microbiol. 293:213-217. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

