Comparison of the genome sequence of the poultry pathogen Bordetella avium with those of B. bronchiseptica, B. pertussis, and B. parapertussis reveals extensive diversity in surface structures associated with host interaction
- PMID: 16885469
- PMCID: PMC1540077
- DOI: 10.1128/JB.01927-05
Comparison of the genome sequence of the poultry pathogen Bordetella avium with those of B. bronchiseptica, B. pertussis, and B. parapertussis reveals extensive diversity in surface structures associated with host interaction
Abstract
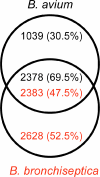
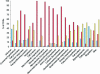
Bordetella avium is a pathogen of poultry and is phylogenetically distinct from Bordetella bronchiseptica, Bordetella pertussis, and Bordetella parapertussis, which are other species in the Bordetella genus that infect mammals. In order to understand the evolutionary relatedness of Bordetella species and further the understanding of pathogenesis, we obtained the complete genome sequence of B. avium strain 197N, a pathogenic strain that has been extensively studied. With 3,732,255 base pairs of DNA and 3,417 predicted coding sequences, it has the smallest genome and gene complement of the sequenced bordetellae. In this study, the presence or absence of previously reported virulence factors from B. avium was confirmed, and the genetic bases for growth characteristics were elucidated. Over 1,100 genes present in B. avium but not in B. bronchiseptica were identified, and most were predicted to encode surface or secreted proteins that are likely to define an organism adapted to the avian rather than the mammalian respiratory tracts. These include genes coding for the synthesis of a polysaccharide capsule, hemagglutinins, a type I secretion system adjacent to two very large genes for secreted proteins, and unique genes for both lipopolysaccharide and fimbrial biogenesis. Three apparently complete prophages are also present. The BvgAS virulence regulatory system appears to have polymorphisms at a poly(C) tract that is involved in phase variation in other bordetellae. A number of putative iron-regulated outer membrane proteins were predicted from the sequence, and this regulation was confirmed experimentally for five of these.
Figures



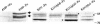
References
-
- Akerley, B. J., P. A. Cotter, and J. F. Miller. 1995. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80:611-620. - PubMed
-
- Allen, A., and D. Maskell. 1996. The identification, cloning and mutagenesis of a genetic locus required for lipopolysaccharide biosynthesis in Bordetella pertussis. Mol. Microbiol. 19:37-52. - PubMed
-
- Allen, A. G., R. M. Thomas, J. T. Cadisch, and D. J. Maskell. 1998. Molecular and functional analysis of the lipopolysaccharide biosynthesis locus wlb from Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Mol. Microbiol. 29:27-38. - PubMed
-
- Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

