Domain characterization of rabbit skeletal muscle myosin light chain kinase
- PMID: 1688558
- PMCID: PMC4325999
Domain characterization of rabbit skeletal muscle myosin light chain kinase
Abstract
Myosin light chain kinase can be divided into three distinct structural domains, an amino-terminal "tail," of unknown function, a central catalytic core and a carboxy-terminal calmodulin-binding regulatory region. We have used a combination of deletion mutagenesis and monoclonal antibody epitope mapping to define these domains more closely. A 2.95-kilobase cDNA has been isolated that includes the entire coding sequence of rabbit skeletal muscle myosin light chain kinase (607 amino acids). This cDNA, expressed in COS cells encoded a Ca2+/calmodulin-dependent myosin light chain kinase with a specific activity similar to that of the enzyme purified from rabbit skeletal muscle. Serial carboxy-terminal deletions of the regulatory and catalytic domains were constructed and expressed in COS cells. The truncated kinases had no detectable myosin light chain kinase activity. Monoclonal antibodies which inhibit the activity of the enzyme competitively with respect to myosin light chain were found to bind between residues 235-319 and 165-173, amino-terminal of the previously defined catalytic core. Thus, residues that are either involved in substrate binding or in close proximity to a light chain binding site may be located more amino-terminal than the previously defined catalytic core.
Figures






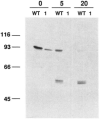

References
-
- Andersson S, Davis DN, Dahlbäck H, Jörnvall H, Russell DW. J. Biol. Chem. 1989;264:8222–8229. - PubMed
-
- Blumenthal DK, Stull JT. Biochemistry. 1980;19:5608–5614. - PubMed
-
- Blumenthal DK, Stull JT. Biochemistry. 1982;21:2386–2391. - PubMed
-
- Edelman AM, Takio K, Blumenthal DK, Hansen RS, Walsh KA, Titani K, Krebs EG. J. Biol. Chem. 1985;260:11275–11285. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

