Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens
- PMID: 16891480
- PMCID: PMC1594621
- DOI: 10.1128/JCM.00542-06
Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens
Abstract
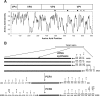
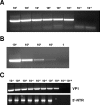
A reverse transcription-seminested PCR (RT-snPCR) assay was developed for the detection and identification of enterovirus (EV) RNA in clinical specimens. Three conserved protein motifs were identified by aligning the VP3 and VP1 sequences of prototype EV strains. Consensus degenerate primers were designed from a conserved VP3 motif and a distal VP1 motif for the first PCR. Consensus-degenerate hybrid oligonucleotide primers were designed from an internal VP1 motif and used with the same distal VP1 motif for the second, seminested PCR step. The primers were designed for broad target specificity and amplified all recognized and proposed EV serotypes and other antigenic variant strains tested. The VP1 RT-snPCR assay was slightly more sensitive than our in-house EV 5' nontranslated region RT-snPCR assay, detecting as few as 10 RNA copies per reaction. As an example application, the VP1 RT-snPCR assay was used to identify EVs in clinical specimens. A product of the expected size was successfully amplified and sequenced from cerebrospinal fluid; serum; stool suspensions; and nasopharyngeal, eye, and rectal swab specimens, allowing unambiguous identification of the infecting virus in all cases. The VP1 sequences derived from the RT-snPCR products allow rapid phylogenetic and molecular epidemiologic analysis of strains circulating during the EV season and comparison with EV sequences from past seasons or from different locations around the world.
Figures

References
-
- Barrios Olivera, J. A., L. Sarmiento Pérez, O. Valdés, P. Mas Lago, and R. Palomera Puentes. 2003. Aplicación de la secuenciación de VP1 a la identificación de enterovirus humanos. Rev. Cubana Med. Trop. 55:133-137. - PubMed
-
- Bolanaki, E., C. Kottaridi, P. Markoulatos, L. Margaritis, and T. Katsorchis. 2005. Nucleotide analysis and phylogenetic study of the homology boundaries of coxsackie A and B viruses. Virus Genes 31:307-320. - PubMed
-
- Caro, V., S. Guillot, F. Delpeyroux, and R. Crainic. 2001. Molecular strategy for ‘serotyping’ of human enteroviruses. J. Gen. Virol. 82:79-91. - PubMed
-
- Casas, I., G. F. Palacios, G. Trallero, D. Cisterna, M. C. Freire, and A. Tenorio. 2001. Molecular characterization of human enteroviruses in clinical samples: comparison between VP2, VP1, and RNA polymerase regions using RT nested PCR assays and direct sequencing of products. J. Med. Virol. 65:138-148. - PubMed
-
- Hamilton, M. S., M. A. Jackson, and D. Abel. 1999. Clinical utility of polymerase chain reaction testing for enteroviral meningitis. Pediatr. Infect. Dis. J. 18:533-537. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

