Real-time quantitative broad-range PCR assay for detection of the 16S rRNA gene followed by sequencing for species identification
- PMID: 16891488
- PMCID: PMC1594602
- DOI: 10.1128/JCM.00112-06
Real-time quantitative broad-range PCR assay for detection of the 16S rRNA gene followed by sequencing for species identification
Abstract
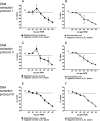
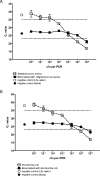
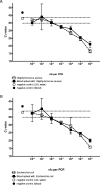
Here we determined the analytical sensitivities of broad-range real-time PCR-based assays employing one of three different genomic DNA extraction protocols in combination with one of three different primer pairs targeting the 16S rRNA gene to detect a panel of 22 bacterial species. DNA extraction protocol III, using lysozyme, lysostaphin, and proteinase K, followed by PCR with the primer pair Bak11W/Bak2, giving amplicons of 796 bp in length, showed the best overall sensitivity, detecting DNA of 82% of the strains investigated at concentrations of < or =10(2) CFU in water per reaction. DNA extraction protocols I and II, using less enzyme treatment, combined with other primer pairs giving shorter amplicons of 466 bp and 342 or 346 bp, respectively, were slightly more sensitive for the detection of gram-negative but less sensitive for the detection of gram-positive bacteria. The obstacle of detecting background DNA in blood samples spiked with bacteria was circumvented by introducing a broad-range hybridization probe, and this preserved the minimal detection limits observed in samples devoid of blood. Finally, sequencing of the amplicons generated using the primer pair Bak11W/Bak2 allowed species identification of the detected bacterial DNA. Thus, broad-spectrum PCR targeting the 16S rRNA gene in the quantitative real-time format can achieve an analytical sensitivity of 1 to 10 CFU per reaction in water, avoid detection of background DNA with the introduction of a broad-range probe, and generate amplicons that allow species identification of the detected bacterial DNA by sequencing. These prerequisites are important for its application to blood-containing patient samples.
Figures

References
-
- Bosshard, P. P., A. Kronenberg, R. Zbinden, C. Ruef, E. C. Bottger, and M. Altwegg. 2003. Etiologic diagnosis of infective endocarditis by broad-range polymerase chain reaction: a 3-year experience. Clin. Infect. Dis. 37:167-172. - PubMed
-
- Böttger, E. C. 1990. Frequent contamination of Taq polymerase with DNA. Clin. Chem. 36:1258-1259. - PubMed
-
- Brun-Buisson, C., M. Fartoukh, E. Lechapt, S. Honore, J. R. Zahar, C. Cerf, and B. Maitre. 2005. Contribution of blinded, protected quantitative specimens to the diagnostic and therapeutic management of ventilator-associated pneumonia. Chest 128:533-544. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

