Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription
- PMID: 16893449
- PMCID: PMC1570494
- DOI: 10.1186/1742-4690-3-48
Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription
Abstract
Background: Human immunodeficiency virus type 1 (HIV-1) is the etiologic agent of acquired immunodeficiency virus (AIDS). Following entry into the host cell, the viral RNA is reverse transcribed into DNA and subsequently integrated into the host genome as a chromatin template. The integrated proviral DNA, along with the specific chromatinized environment in which integration takes place allows for the coordinated regulation of viral transcription and replication. While the specific roles of and interplay between viral and host proteins have not been fully elucidated, numerous reports indicate that HIV-1 retains the ability for self-regulation via the pleiotropic effects of its viral proteins. Though viral transcription is fully dependent upon host cellular factors and the state of host activation, recent findings indicate a complex interplay between viral proteins and host transcription regulatory machineries including histone deacetylases (HDACs), histone acetyltransferases (HATs), cyclin dependent kinases (CDKs), and histone methyltransferases (HMTs).
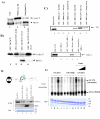
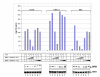
Results: Here, we describe the effect of Tat activated transcription at the G1/S border of the cell cycle and analyze the interaction of modified Tat with the chromatin remodeling complex, SWI/SNF. HIV-1 LTR DNA reconstituted into nucleosomes can be activated in vitro using various Tat expressing extracts. Optimally activated transcription was observed at the G1/S border of the cell cycle both in vitro and in vivo, where chromatin remodeling complex, SWI/SNF, was present on the immobilized LTR DNA. Using a number of in vitro binding as well as in vivo chromatin immunoprecipitation (ChIP) assays, we detected the presence of both BRG1 and acetylated Tat in the same complex. Finally, we demonstrate that activated transcription resulted in partial or complete removal of the nucleosome from the start site of the LTR as evidenced by a restriction enzyme accessibility assay.
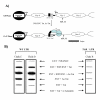
Conclusion: We propose a model where unmodified Tat is involved in binding to the CBP/p300 and cdk9/cyclin T1 complexes facilitating transcription initiation. Acetylated Tat dissociates from the TAR RNA structure and recruits bromodomain-binding chromatin modifying complexes such as p/CAF and SWI/SNF to possibly facilitate transcription elongation.
Figures
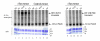




References
-
- Fauci AS. Host factors in the pathogenesis of HIV disease. Antibiot Chemother. 1996;48:4–12. - PubMed
-
- Coull JJ, Romerio F, Sun JM, Volker JL, Galvin KM, Davie JR, Shi Y, Hansen U, Margolis DM. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J Virol. 2000;74:6790–6799. doi: 10.1128/JVI.74.15.6790-6799.2000. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

