Global DNA hypermethylation-associated cancer chemotherapy resistance and its reversion with the demethylating agent hydralazine
- PMID: 16893460
- PMCID: PMC1563479
- DOI: 10.1186/1479-5876-4-32
Global DNA hypermethylation-associated cancer chemotherapy resistance and its reversion with the demethylating agent hydralazine
Abstract
Background: The development of resistance to cytotoxic chemotherapy continues to be a major obstacle for successful anticancer therapy. It has been shown that cells exposed to toxic concentrations of commonly used cancer chemotherapy agents develop DNA hypermethylation. Hence, demethylating agents could play a role in overcoming drug resistance.
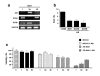
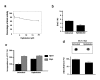
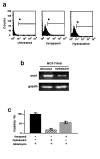
Methods: MCF-7 cells were rendered adriamycin-resistant by weekly treatment with adriamycin. Wild-type and the resulting MCF-7/Adr cells were analyzed for global DNA methylation. DNA methyltransferase activity and DNA methyltransferase (dnmt) gene expression were also determined. MCF-7/Adr cells were then subjected to antisense targeting of dnmt1, -3a, and -b genes and to treatment with the DNA methylation inhibitor hydralazine to investigate whether DNA demethylation restores sensitivity to adriamycin.
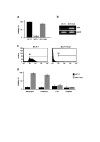
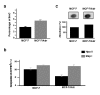
Results: MCF-7/Adr cells exhibited the multi-drug resistant phenotype as demonstrated by adriamycin resistance, mdr1 gene over-expression, decreased intracellular accumulation of adriamycin, and cross-resistance to paclitaxel. The mdr phenotype was accompanied by global DNA hypermethylation, over-expression of dnmt genes, and increased DNA methyltransferase activity as compared with wild-type MCF-7 cells. DNA demethylation through antisense targeting of dnmts or hydralazine restored adriamycin sensitivity of MCF-7/Adr cells to a greater extent than verapamil, a known inhibitor of mdr protein, suggesting that DNA demethylation interferes with the epigenetic reprogramming that participates in the drug-resistant phenotype.
Conclusion: We provide evidence that DNA hypermethylation is at least partly responsible for development of the multidrug-resistant phenotype in the MCF-7/Adr model and that hydralazine, a known DNA demethylating agent, can revert the resistant phenotype.
Figures

Similar articles
-
Reversing adriamycin resistance of human breast cancer cells by hyperthermia combined with Interferon alpha and Verapamil.J Exp Clin Cancer Res. 2007 Jun;26(2):201-7. J Exp Clin Cancer Res. 2007. PMID: 17725099
-
Reversal effect of isotetrandrine, an isoquinoline alkaloid extracted from Caulis Mahoniae, on P-glycoprotein-mediated doxorubicin-resistance in human breast cancer (MCF-7/DOX) cells.Yao Xue Xue Bao. 2008 May;43(5):461-6. Yao Xue Xue Bao. 2008. PMID: 18717331
-
Effects of multidrug resistance, antisense RNA on the chemosensitivity of hepatocellular carcinoma cells.Hepatobiliary Pancreat Dis Int. 2006 Nov;5(4):552-9. Hepatobiliary Pancreat Dis Int. 2006. PMID: 17085341
-
Thermosensitivity and thermotolerance in the adriamycin-resistant strain of Ehrlich ascites tumor cells.Anticancer Res. 1996 Sep-Oct;16(5A):2569-73. Anticancer Res. 1996. PMID: 8917353
-
Targeting cellular memory to reprogram the epigenome, restore potential, and improve somatic cell nuclear transfer.Anim Reprod Sci. 2007 Mar;98(1-2):129-46. doi: 10.1016/j.anireprosci.2006.10.019. Epub 2006 Oct 21. Anim Reprod Sci. 2007. PMID: 17166676 Review.
Cited by
-
Regulation of demethylation and re-expression of RASSF1A gene in gastric cancer cell lines by combined treatment of 5-Aza-CdR and NaB.World J Gastroenterol. 2008 Jan 28;14(4):595-600. doi: 10.3748/wjg.14.595. World J Gastroenterol. 2008. PMID: 18203293 Free PMC article.
-
Low-dose hydralazine during gestation reduces renal fibrosis in rodent offspring exposed to maternal high fat diet.PLoS One. 2021 Mar 18;16(3):e0248854. doi: 10.1371/journal.pone.0248854. eCollection 2021. PLoS One. 2021. PMID: 33735324 Free PMC article.
-
DNA methyltransferase expression in triple-negative breast cancer predicts sensitivity to decitabine.J Clin Invest. 2018 Jun 1;128(6):2376-2388. doi: 10.1172/JCI97924. Epub 2018 Apr 30. J Clin Invest. 2018. PMID: 29708513 Free PMC article.
-
The inhibition of UBC13 expression and blockage of the DNMT1-CHFR-Aurora A pathway contribute to paclitaxel resistance in ovarian cancer.Cell Death Dis. 2018 Jan 24;9(2):93. doi: 10.1038/s41419-017-0137-x. Cell Death Dis. 2018. PMID: 29367628 Free PMC article.
-
ERα propelled aberrant global DNA hypermethylation by activating the DNMT1 gene to enhance anticancer drug resistance in human breast cancer cells.Oncotarget. 2016 Apr 12;7(15):20966-80. doi: 10.18632/oncotarget.8038. Oncotarget. 2016. PMID: 26980709 Free PMC article.
References
-
- Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

