Characterization of the mrgRS locus of the opportunistic pathogen Burkholderia pseudomallei: temperature regulates the expression of a two-component signal transduction system
- PMID: 16893462
- PMCID: PMC1557856
- DOI: 10.1186/1471-2180-6-70
Characterization of the mrgRS locus of the opportunistic pathogen Burkholderia pseudomallei: temperature regulates the expression of a two-component signal transduction system
Abstract
Background: Burkholderia pseudomallei is a saprophyte in tropical environments and an opportunistic human pathogen. This versatility requires a sensing mechanism that allows the bacterium to respond rapidly to altered environmental conditions. We characterized a two-component signal transduction locus from B. pseudomallei 204, mrgR and mrgS, encoding products with extensive homology with response regulators and histidine protein kinases of Escherichia coli, Bordetella pertussis, and Vibrio cholerae.
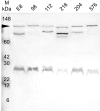
Results: The locus was present and expressed in a variety of B. pseudomallei human and environmental isolates but was absent from other Burkholderia species, B. cepacia, B. cocovenenans, B. plantarii, B. thailandensis, B. vandii, and B. vietnamiensis. A 2128 bp sequence, including the full response regulator mrgR, but not the sensor kinase mrgS, was present in the B. mallei genome. Restriction fragment length polymorphism downstream from mrgRS showed two distinct groups were present among B. pseudomallei isolates. Our analysis of the open reading frames in this region of the genome revealed that transposase and bacteriophage activity may help explain this variation. MrgR and MrgS proteins were expressed in B. pseudomallei 204 cultured at different pH, salinity and temperatures and the expression was substantially reduced at 25 degrees C compared with 37 degrees C or 42 degrees C but was mostly unaffected by pH or salinity, although at 25 degrees C and 0.15% NaCl a small increase in MrgR expression was observed at pH 5. MrgR was recognized by antibodies in convalescent sera pooled from melioidosis patients.
Conclusion: The results suggest that mrgRS regulates an adaptive response to temperature that may be essential for pathogenesis, particularly during the initial phases of infection. B. pseudomallei and B. mallei are very closely related species that differ in their capacity to adapt to changing environmental conditions. Modifications in this region of the genome may assist our understanding of the reasons for this difference.
Figures
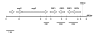






Similar articles
-
Single gene target bacterial identification. groEL gene sequencing for discriminating clinical isolates of Burkholderia pseudomallei and Burkholderia thailandensis.Diagn Microbiol Infect Dis. 2002 Oct;44(2):143-9. doi: 10.1016/s0732-8893(02)00439-x. Diagn Microbiol Infect Dis. 2002. PMID: 12458120
-
Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei.J Clin Microbiol. 2003 May;41(5):2068-79. doi: 10.1128/JCM.41.5.2068-2079.2003. J Clin Microbiol. 2003. PMID: 12734250 Free PMC article.
-
Identification of Burkholderia mallei and Burkholderia pseudomallei adhesins for human respiratory epithelial cells.BMC Microbiol. 2010 Sep 28;10:250. doi: 10.1186/1471-2180-10-250. BMC Microbiol. 2010. PMID: 20920184 Free PMC article.
-
Secondary metabolites from the Burkholderia pseudomallei complex: structure, ecology, and evolution.J Ind Microbiol Biotechnol. 2020 Oct;47(9-10):877-887. doi: 10.1007/s10295-020-02317-0. Epub 2020 Oct 14. J Ind Microbiol Biotechnol. 2020. PMID: 33052546 Free PMC article. Review.
-
PCR-based Methodologies Used to Detect and Differentiate the Burkholderia pseudomallei complex: B. pseudomallei, B. mallei, and B. thailandensis.Curr Issues Mol Biol. 2014;16:23-54. Epub 2013 Aug 22. Curr Issues Mol Biol. 2014. PMID: 23969318 Review.
Cited by
-
Melioidosis in Birds and Burkholderia pseudomallei Dispersal, Australia.Emerg Infect Dis. 2011 Jul;17(7):1310-2. doi: 10.3201/eid1707.100707. Emerg Infect Dis. 2011. PMID: 21762599 Free PMC article. No abstract available.
-
Enzymatic and molecular characterisation of leucine aminopeptidase of Burkholderia pseudomallei.BMC Microbiol. 2013 May 17;13:110. doi: 10.1186/1471-2180-13-110. BMC Microbiol. 2013. PMID: 23682954 Free PMC article.
-
Regulation of Virulence by Two-Component Systems in Pathogenic Burkholderia.Infect Immun. 2020 Jun 22;88(7):e00927-19. doi: 10.1128/IAI.00927-19. Print 2020 Jun 22. Infect Immun. 2020. PMID: 32284365 Free PMC article. Review.
-
Perturbation of the two-component signal transduction system, BprRS, results in attenuated virulence and motility defects in Burkholderia pseudomallei.BMC Genomics. 2016 May 4;17:331. doi: 10.1186/s12864-016-2668-4. BMC Genomics. 2016. PMID: 27147217 Free PMC article.
References
-
- Pruksachartvuthi S, Aswapokee N, Thankerngpol K. Survival of Pseudomonas pseudomallei in human phagocytes. J Med Microbiol. 1990;31:109–114. - PubMed
-
- Everett ED, Nelson RA. Pulmonary melioidosis. Observations in thirty-nine cases. Am Rev Resp Dis. 1975;112:331–340. - PubMed
-
- So SY, Chau PY, Leung YK, Lam WK, Yu DYC. Successful treatment of melioidosis caused by a multiresistant strain in an immunocompromised host with third generation cepharosporins. Am Rev Resp Dis. 1983;127:650–654. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

