Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast
- PMID: 16893973
- PMCID: PMC2064256
- DOI: 10.1083/jcb.200605074
Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast
Abstract
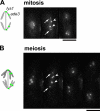
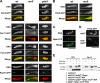
The meiotic cohesin Rec8 is required for the stepwise segregation of chromosomes during the two rounds of meiotic division. By directly measuring chromosome compaction in living cells of the fission yeast Schizosaccharomyces pombe, we found an additional role for the meiotic cohesin in the compaction of chromosomes during meiotic prophase. In the absence of Rec8, chromosomes were decompacted relative to those of wild-type cells. Conversely, loss of the cohesin-associated protein Pds5 resulted in hypercompaction. Although this hypercompaction requires Rec8, binding of Rec8 to chromatin was reduced in the absence of Pds5, indicating that Pds5 promotes chromosome association of Rec8. To explain these observations, we propose that meiotic prophase chromosomes are organized as chromatin loops emanating from a Rec8-containing axis: the absence of Rec8 disrupts the axis, resulting in disorganized chromosomes, whereas reduced Rec8 loading results in a longitudinally compacted axis with fewer attachment points and longer chromatin loops.
Figures






References
-
- Bahler, J., J.Q. Wu, M.S. Longtine, N.G. Shah, A. McKenzie III, A.B. Steever, A. Wach, P. Philippsen, and J.R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951. - PubMed
-
- Bannister, L.A., L.G. Reinholdt, R.J. Munroe, and J.C. Schimenti. 2004. Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis. 40:184–194. - PubMed
-
- Cervantes, M.D., J.A. Farah, and G.R. Smith. 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell. 5:883–888. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

