The yeast ubiquitin ligase SCFMet30: connecting environmental and intracellular conditions to cell division
- PMID: 16895602
- PMCID: PMC1579207
- DOI: 10.1186/1747-1028-1-16
The yeast ubiquitin ligase SCFMet30: connecting environmental and intracellular conditions to cell division
Abstract
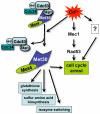
Ubiquitination regulates a host of cellular processes and is well known for its role in progression through the cell division cycle. In budding yeast, cadmium and arsenic stress, the availability of sulfur containing amino acids, and the intracellular concentration of S-adenosylmethionine are linked to cell cycle regulation through the ubiquitin ligase SCFMet30. Regulation is achieved by ubiquitination of the transcription factor Met4. Met4 activity is controlled by a regulatory K48-linked ubiquitin chain that is synthesized by Cdc34/SCFMet30. A ubiquitin-interacting-motif (UIM) present in Met4 prevents degradation of ubiquitinated Met4 allowing the ubiquitin chain to function as a reversible switch of Met4 activity. Here we discuss mechanisms of Met4 and SCFMet30 regulation in response to intracellular and environmental conditions, and describe the integration of these signals with cell cycle control.
Figures


Similar articles
-
The yeast ubiquitin ligase SCFMet30 regulates heavy metal response.Mol Biol Cell. 2005 Apr;16(4):1872-82. doi: 10.1091/mbc.e04-12-1130. Epub 2005 Feb 2. Mol Biol Cell. 2005. PMID: 15689486 Free PMC article.
-
Destabilization of binding to cofactors and SCFMet30 is the rate-limiting regulatory step in degradation of polyubiquitinated Met4.Mol Cell. 2006 Dec 8;24(5):689-699. doi: 10.1016/j.molcel.2006.10.028. Mol Cell. 2006. Retraction in: Mol Cell. 2018 Apr 19;70(2):381. doi: 10.1016/j.molcel.2018.03.002. PMID: 17157252 Retracted.
-
The emerging regulatory potential of SCFMet30 -mediated polyubiquitination and proteolysis of the Met4 transcriptional activator.Cell Div. 2008 Jul 25;3:11. doi: 10.1186/1747-1028-3-11. Cell Div. 2008. PMID: 18655704 Free PMC article.
-
Role of the ubiquitin-proteasome system in brain ischemia: friend or foe?Prog Neurobiol. 2014 Jan;112:50-69. doi: 10.1016/j.pneurobio.2013.10.003. Epub 2013 Oct 22. Prog Neurobiol. 2014. PMID: 24157661 Review.
-
[Ubiquitin-proteasome pathway and virus infection].Sheng Wu Gong Cheng Xue Bao. 2004 Mar;20(2):151-6. Sheng Wu Gong Cheng Xue Bao. 2004. PMID: 15969100 Review. Chinese.
Cited by
-
A transcriptional activator is part of an SCF ubiquitin ligase to control degradation of its cofactors.Mol Cell. 2010 Dec 22;40(6):954-64. doi: 10.1016/j.molcel.2010.11.018. Mol Cell. 2010. PMID: 21172660 Free PMC article.
-
Cdc48 cofactor Shp1 regulates signal-induced SCFMet30 disassembly.Proc Natl Acad Sci U S A. 2020 Sep 1;117(35):21319-21327. doi: 10.1073/pnas.1922891117. Epub 2020 Aug 18. Proc Natl Acad Sci U S A. 2020. PMID: 32817489 Free PMC article.
-
Cadmium binding by the F-box domain induces p97-mediated SCF complex disassembly to activate stress response programs.Nat Commun. 2024 May 8;15(1):3894. doi: 10.1038/s41467-024-48184-6. Nat Commun. 2024. PMID: 38719837 Free PMC article.
-
Modulation of the complex regulatory network for methionine biosynthesis in fungi.Genetics. 2021 Feb 9;217(2):iyaa049. doi: 10.1093/genetics/iyaa049. Genetics. 2021. PMID: 33724418 Free PMC article.
-
Distinct Stress Regulators in the CRL Family: Emerging Roles of F-Box Proteins: Cullin-RING Ligases and Stress-Sensing.Bioessays. 2025 May;47(5):e202400249. doi: 10.1002/bies.202400249. Epub 2025 Mar 16. Bioessays. 2025. PMID: 40091294 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

