Glycoproteomic probes for fluorescent imaging of fucosylated glycans in vivo
- PMID: 16895981
- PMCID: PMC1567886
- DOI: 10.1073/pnas.0605418103
Glycoproteomic probes for fluorescent imaging of fucosylated glycans in vivo
Abstract
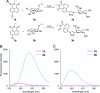
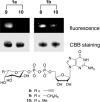
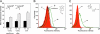
Glycomics is emerging as a new field for the biology of complex glycoproteins and glycoconjugates. The lack of versatile glycan-labeling methods has presented a major obstacle to visualizing at the cellular level and studying glycoconjugates. To address this issue, we developed a fluorescent labeling technique based on the Cu(I)-catalyzed [3 + 2] cycloaddition, or click chemistry, which allows rapid, versatile, and specific covalent labeling of cellular glycans bearing azide groups. The method entails generating a fluorescent probe from a nonfluorescent precursor, 4-ethynyl-N-ethyl-1,8-naphthalimide, by clicking the fluorescent trigger, the alkyne at the 4 position, with an azido-modified sugar. Using this click-activated fluorescent probe, we demonstrate incorporation of an azido-containing fucose analog into glycoproteins via the fucose salvage pathway. Distinct fluorescent signals were observed by flow cytometry when cells treated with 6-azidofucose were labeled with the click-activated fluorogenic probe or biotinylated alkyne. The intracellular localization of fucosylated glycoconjugates was visualized by using fluorescence microscopy. This technique will allow dynamic imaging of cellular fucosylation and facilitate studies of fucosylated glycoproteins and glycolipids.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
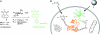



Similar articles
-
Differential Labeling of Glycoproteins with Alkynyl Fucose Analogs.Int J Mol Sci. 2020 Aug 20;21(17):6007. doi: 10.3390/ijms21176007. Int J Mol Sci. 2020. PMID: 32825463 Free PMC article.
-
Alkynyl sugar analogs for the labeling and visualization of glycoconjugates in cells.Proc Natl Acad Sci U S A. 2007 Feb 20;104(8):2614-9. doi: 10.1073/pnas.0611307104. Epub 2007 Feb 12. Proc Natl Acad Sci U S A. 2007. PMID: 17296930 Free PMC article.
-
Imaging glycans in zebrafish embryos by metabolic labeling and bioorthogonal click chemistry.J Vis Exp. 2011 Jun 6;(52):2686. doi: 10.3791/2686. J Vis Exp. 2011. PMID: 21673647 Free PMC article.
-
Direct Visualization of Live Zebrafish Glycans via Single-Step Metabolic Labeling with Fluorophore-Tagged Nucleotide Sugars.Angew Chem Int Ed Engl. 2019 Oct 1;58(40):14327-14333. doi: 10.1002/anie.201907410. Epub 2019 Aug 28. Angew Chem Int Ed Engl. 2019. PMID: 31295389 Free PMC article. Review.
-
Metabolic utilization and remodeling of glycan biosynthesis using fucose analogs.Biochim Biophys Acta Gen Subj. 2022 Dec;1866(12):130243. doi: 10.1016/j.bbagen.2022.130243. Epub 2022 Sep 7. Biochim Biophys Acta Gen Subj. 2022. PMID: 36087787 Review.
Cited by
-
Highly selective fluorescent and colorimetric probe for live-cell monitoring of sulphide based on bioorthogonal reaction.Sci Rep. 2015 Mar 11;5:8969. doi: 10.1038/srep08969. Sci Rep. 2015. PMID: 25759082 Free PMC article.
-
Small molecule probes for plant cell wall polysaccharide imaging.Front Plant Sci. 2012 May 11;3:89. doi: 10.3389/fpls.2012.00089. eCollection 2012. Front Plant Sci. 2012. PMID: 22639673 Free PMC article.
-
Development of orally active inhibitors of protein and cellular fucosylation.Proc Natl Acad Sci U S A. 2013 Apr 2;110(14):5404-9. doi: 10.1073/pnas.1222263110. Epub 2013 Mar 14. Proc Natl Acad Sci U S A. 2013. PMID: 23493549 Free PMC article.
-
6-alkynyl fucose is a bioorthogonal analog for O-fucosylation of epidermal growth factor-like repeats and thrombospondin type-1 repeats by protein O-fucosyltransferases 1 and 2.Glycobiology. 2013 Feb;23(2):188-98. doi: 10.1093/glycob/cws140. Epub 2012 Oct 8. Glycobiology. 2013. PMID: 23045360 Free PMC article.
-
Differential Labeling of Glycoproteins with Alkynyl Fucose Analogs.Int J Mol Sci. 2020 Aug 20;21(17):6007. doi: 10.3390/ijms21176007. Int J Mol Sci. 2020. PMID: 32825463 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

