Antagonism between two mechanisms of antifungal drug resistance
- PMID: 16896209
- PMCID: PMC1539145
- DOI: 10.1128/EC.00048-06
Antagonism between two mechanisms of antifungal drug resistance
Abstract
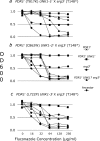
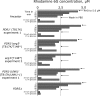
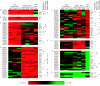
This study tested for interaction between two independently evolved mechanisms of fluconazole resistance in Saccharomyces cerevisiae. One set of strains was from a 400-generation evolution experiment, during which the concentration of fluconazole was increased from 16 to 256 microg/ml in four increments. At 100 generations, populations became fixed for resistance mutations in either of two transcriptional regulators, PDR1 or PDR3. At 400 generations, replicate populations became fixed for another resistance mutation in UNK1, an unmapped gene further increasing resistance. Another genotype used in this study came from a population placed initially in 128 microg/ml of fluconazole; this environment selects for resistance through loss of function at ERG3, resulting in altered sterol metabolism. Mutant strains carrying PDR1(r) or PDR3(r) were crossed with the erg3(r) mutant strain, and the doubly mutant, haploid offspring were identified. The double-mutant strains grew less well than the parent strains at all concentrations of fluconazole tested. In genome-wide assays of gene expression, several ABC transporter genes that were overexpressed in one parent and several ERG genes that were overexpressed in the other parent were also overexpressed in the double mutants. Of the 43 genes that were consistently overexpressed in the PDR1(r) parents at generation 100, however, 31 were not consistently overexpressed in the double mutants. Of these 31 genes, 30 were also not consistently overexpressed after a further 300 generations of evolution in the PDR1(r) parent populations. The two independently evolved mechanisms of fluconazole resistance are strongly antagonistic to one another.
Figures


Similar articles
-
The ATP-binding cassette multidrug transporter Snq2 of Saccharomyces cerevisiae: a novel target for the transcription factors Pdr1 and Pdr3.Mol Microbiol. 1996 Apr;20(1):109-17. doi: 10.1111/j.1365-2958.1996.tb02493.x. Mol Microbiol. 1996. PMID: 8861209
-
Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies.Mol Microbiol. 2006 Aug;61(3):704-22. doi: 10.1111/j.1365-2958.2006.05235.x. Epub 2006 Jun 27. Mol Microbiol. 2006. PMID: 16803598
-
Haploidy, diploidy and evolution of antifungal drug resistance in Saccharomyces cerevisiae.Genetics. 2004 Dec;168(4):1915-23. doi: 10.1534/genetics.104.033266. Epub 2004 Sep 15. Genetics. 2004. PMID: 15371350 Free PMC article.
-
[What's new in the molecular basis of antifungal drug resistance in pathogenic yeasts].Nihon Rinsho. 2008 Dec;66(12):2273-8. Nihon Rinsho. 2008. PMID: 19069091 Review. Japanese.
-
Unveiling the transcriptional control of pleiotropic drug resistance in Saccharomyces cerevisiae: Contributions of André Goffeau and his group.Yeast. 2019 Apr;36(4):195-200. doi: 10.1002/yea.3354. Epub 2018 Oct 3. Yeast. 2019. PMID: 30194700 Free PMC article. Review.
Cited by
-
Discovery of an orally active nitrothiophene-based antitrypanosomal agent.Eur J Med Chem. 2024 Jan 5;263:115954. doi: 10.1016/j.ejmech.2023.115954. Epub 2023 Nov 15. Eur J Med Chem. 2024. PMID: 37984297 Free PMC article.
-
Gene expression and evolution of antifungal drug resistance.Antimicrob Agents Chemother. 2009 May;53(5):1931-6. doi: 10.1128/AAC.01315-08. Epub 2009 Mar 9. Antimicrob Agents Chemother. 2009. PMID: 19273689 Free PMC article.
-
Vinyl sulfone-based inhibitors of trypanosomal cysteine protease rhodesain with improved antitrypanosomal activities.Bioorg Med Chem Lett. 2020 Jul 15;30(14):127217. doi: 10.1016/j.bmcl.2020.127217. Epub 2020 Apr 28. Bioorg Med Chem Lett. 2020. PMID: 32527539 Free PMC article.
-
Suppressive drug combinations and their potential to combat antibiotic resistance.J Antibiot (Tokyo). 2017 Nov;70(11):1033-1042. doi: 10.1038/ja.2017.102. Epub 2017 Sep 6. J Antibiot (Tokyo). 2017. PMID: 28874848 Free PMC article. Review.
-
Genetic and genomic architecture of the evolution of resistance to antifungal drug combinations.PLoS Genet. 2013 Apr;9(4):e1003390. doi: 10.1371/journal.pgen.1003390. Epub 2013 Apr 4. PLoS Genet. 2013. PMID: 23593013 Free PMC article.
References
-
- Agarwal, A. K., P. D. Rogers, S. R. Baerson, M. R. Jacob, K. S. Barker, J. D. Cleary, L. A. Walker, D. G. Nagle, and A. M. Clark. 2003. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 278:34998-35015. - PubMed
-
- Anderson, J. B. 2005. Evolution of antifungal-drug resistance: mechanisms and pathogen fitness. Nat. Rev. Microbiol. 3:547-556. - PubMed
-
- Cowen, L. E., and S. Lindquist. 2005. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science 309:2185-2189. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

