Genomic view of the evolution of the complement system
- PMID: 16896831
- PMCID: PMC2480602
- DOI: 10.1007/s00251-006-0142-1
Genomic view of the evolution of the complement system
Abstract
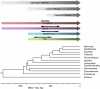
The recent accumulation of genomic information of many representative animals has made it possible to trace the evolution of the complement system based on the presence or absence of each complement gene in the analyzed genomes. Genome information from a few mammals, chicken, clawed frog, a few bony fish, sea squirt, fruit fly, nematoda and sea anemone indicate that bony fish and higher vertebrates share practically the same set of complement genes. This suggests that most of the gene duplications that played an essential role in establishing the mammalian complement system had occurred by the time of the teleost/mammalian divergence around 500 million years ago (MYA). Members of most complement gene families are also present in ascidians, although they do not show a one-to-one correspondence to their counterparts in higher vertebrates, indicating that the gene duplications of each gene family occurred independently in vertebrates and ascidians. The C3 and factor B genes, but probably not the other complement genes, are present in the genome of the cnidaria and some protostomes, indicating that the origin of the central part of the complement system was established more than 1,000 MYA.
Figures



References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor Miklos GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C et al (2000) The genome sequence of Drosophila melanogaster. Science 287:2185–2195 - DOI - PubMed
-
- Al-Sharif WZ, Sunyer JO, Lambris JD, Smith LC (1998) Sea urchin coelomocytes specifically express a homologue of the complement component C3. J Immunol 160:2983–2997 - PubMed
-
- Azumi K, De Santis R, De Tomaso A, Rigoutsos I, Yoshizaki F, Pinto M, Marino A, Shida K, Ikeda M, Ikeda M, Arai M, Inoue Y, Shimizu T, Satoh N, Rokhsar D, Du Pasquier L, Kasahara M, Satake M, Nonaka M (2003) Genomic analysis of immunity in a Urochordate and the emergence of the vertebrate immune system: “waiting for Godot.” Immunogenetics 55(8):570–581 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

