Gene and protein nomenclature in public databases
- PMID: 16899134
- PMCID: PMC1560172
- DOI: 10.1186/1471-2105-7-372
Gene and protein nomenclature in public databases
Abstract
Background: Frequently, several alternative names are in use for biological objects such as genes and proteins. Applications like manual literature search, automated text-mining, named entity identification, gene/protein annotation, and linking of knowledge from different information sources require the knowledge of all used names referring to a given gene or protein. Various organism-specific or general public databases aim at organizing knowledge about genes and proteins. These databases can be used for deriving gene and protein name dictionaries. So far, little is known about the differences between databases in terms of size, ambiguities and overlap.
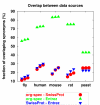
Results: We compiled five gene and protein name dictionaries for each of the five model organisms (yeast, fly, mouse, rat, and human) from different organism-specific and general public databases. We analyzed the degree of ambiguity of gene and protein names within and between dictionaries, to a lexicon of common English words and domain-related non-gene terms, and we compared different data sources in terms of size of extracted dictionaries and overlap of synonyms between those. The study shows that the number of genes/proteins and synonyms covered in individual databases varies significantly for a given organism, and that the degree of ambiguity of synonyms varies significantly between different organisms. Furthermore, it shows that, despite considerable efforts of co-curation, the overlap of synonyms in different data sources is rather moderate and that the degree of ambiguity of gene names with common English words and domain-related non-gene terms varies depending on the considered organism.
Conclusion: In conclusion, these results indicate that the combination of data contained in different databases allows the generation of gene and protein name dictionaries that contain significantly more used names than dictionaries obtained from individual data sources. Furthermore, curation of combined dictionaries considerably increases size and decreases ambiguity. The entries of the curated synonym dictionary are available for manual querying, editing, and PubMed- or Google-search via the ProThesaurus-wiki. For automated querying via custom software, we offer a web service and an exemplary client application.
Figures



References
-
- Balakrishnan R, Christie KR, Costanzo MC, Dolinski K, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hong EL, Nash R, Oughtred R, Skrzypek M, Theesfeld CL, Binkley G, Dong Q, Lane C, Sethuraman A, Weng S, Botstein D, Cherry JM. Fungal BLAST and Model Organism BLASTP Best Hits: new comparison resources at the Saccharomyces Genome Database (SGD) Nucleic Acids Res. 2005:D374–7. - PMC - PubMed
-
- Drysdale RA, Crosby MA, Gelbart W, Campbell K, Emmert D, Matthews B, Russo S, Schroeder A, Smutniak F, Zhang P, Zhou P, Zytkovicz M, Ashburner M, de Grey A, Foulger R, Millburn G, Sutherland D, Yamada C, Kaufman T, Matthews K, DeAngelo A, Cook RK, Gilbert D, Goodman J, Grumbling G, Sheth H, Strelets V, Rubin G, Gibson M, Harris N, Lewis S, Misra S, Shu SQ. FlyBase: genes and gene models. Nucleic Acids Res. 2005:D390–5. - PMC - PubMed
-
- Eppig JT, Bult CJ, Kadin JA, Richardson JE, Blake JA, Anagnostopoulos A, Baldarelli RM, Baya M, Beal JS, Bello SM, Boddy WJ, Bradt DW, Burkart DL, Butler NE, Campbell J, Cassell MA, Corbani LE, Cousins SL, Dahmen DJ, Dene H, Diehl AD, Drabkin HJ, Frazer KS, Frost P, Glass LH, Goldsmith CW, Grant PL, Lennon-Pierce M, Lewis J, Lu I, Maltais LJ, McAndrews-Hill M, McClellan L, Miers DB, Miller LA, Ni L, Ormsby JE, Qi D, Reddy TB, Reed DJ, Richards-Smith B, Shaw DR, Sinclair R, Smith CL, Szauter P, Walker MB, Walton DO, Washburn LL, Witham IT, Zhu Y. The Mouse Genome Database (MGD): from genes to mice – a community resource for mouse biology. Nucleic Acids Res. 2005:D471–5. - PMC - PubMed
-
- de la Cruz N, Bromberg S, Pasko D, Shimoyama M, Twigger S, Chen J, Chen CF, Fan C, Foote C, Gopinath GR, Harris G, Hughes A, Ji Y, Jin W, Li D, Mathis J, Nenasheva N, Nie J, Nigam R, Petri V, Reilly D, Wang W, Wu W, Zuniga-Meyer A, Zhao L, Kwitek A, Tonellato P, Jacob H. The Rat Genome Database (RGD): developments towards a phenome database. Nucleic Acids Res. 2005:D485–91. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

