Gamma-tubulin is required for proper recruitment and assembly of Kar9-Bim1 complexes in budding yeast
- PMID: 16899509
- PMCID: PMC1635365
- DOI: 10.1091/mbc.e06-03-0245
Gamma-tubulin is required for proper recruitment and assembly of Kar9-Bim1 complexes in budding yeast
Abstract
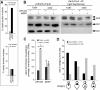
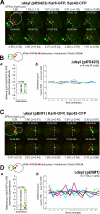
Microtubule plus-end-interacting proteins (+TIPs) promote the dynamic interactions between the plus ends (+ends) of astral microtubules and cortical actin that are required for preanaphase spindle positioning. Paradoxically, +TIPs such as the EB1 orthologue Bim1 and Kar9 also associate with spindle pole bodies (SPBs), the centrosome equivalent in budding yeast. Here, we show that deletion of four C-terminal residues of the budding yeast gamma-tubulin Tub4 (tub4-delta dsyl) perturbs Bim1 and Kar9 localization to SPBs and Kar9-dependent spindle positioning. Surprisingly, we find Kar9 localizes to microtubule +ends in tub4-delta dsyl cells, but these microtubules fail to position the spindle when targeted to the bud. Using cofluorescence and coaffinity purification, we show Kar9 complexes in tub4-delta dsyl cells contain reduced levels of Bim1. Astral microtubule dynamics is suppressed in tub4-delta dsyl cells, but it are restored by deletion of Kar9. Moreover, Myo2- and F-actin-dependent dwelling of Kar9 in the bud is observed in tub4-delta dsyl cells, suggesting defective Kar9 complexes tether microtubule +ends to the cortex. Overproduction of Bim1, but not Kar9, restores Kar9-dependent spindle positioning in the tub4-delta dsyl mutant, reduces cortical dwelling, and promotes Bim1-Kar9 interactions. We propose that SPBs, via the tail of Tub4, promote the assembly of functional +TIP complexes before their deployment to microtubule +ends.
Figures



Similar articles
-
Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables.J Cell Biol. 2003 May 12;161(3):483-8. doi: 10.1083/jcb.200302030. J Cell Biol. 2003. PMID: 12743102 Free PMC article.
-
Cdk1-Clb4 controls the interaction of astral microtubule plus ends with subdomains of the daughter cell cortex.Genes Dev. 2004 Jul 15;18(14):1709-24. doi: 10.1101/gad.298704. Genes Dev. 2004. PMID: 15256500 Free PMC article.
-
Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions.EMBO J. 2003 Feb 3;22(3):438-49. doi: 10.1093/emboj/cdg063. EMBO J. 2003. PMID: 12554645 Free PMC article.
-
Kar9 asymmetrical loading on spindle poles mediates proper spindle alignment in budding yeast.Dev Cell. 2003 Mar;4(3):289-90. doi: 10.1016/s1534-5807(03)00065-0. Dev Cell. 2003. PMID: 12636909 Review.
-
Astral microtubule asymmetry provides directional cues for spindle positioning in budding yeast.Exp Cell Res. 2012 Jul 15;318(12):1400-6. doi: 10.1016/j.yexcr.2012.04.006. Epub 2012 Apr 19. Exp Cell Res. 2012. PMID: 22542856 Free PMC article. Review.
Cited by
-
Actin- and microtubule-based motors contribute to clathrin-independent endocytosis in yeast.Mol Biol Cell. 2023 Nov 1;34(12):ar117. doi: 10.1091/mbc.E23-05-0164. Epub 2023 Aug 30. Mol Biol Cell. 2023. PMID: 37647159 Free PMC article.
-
Concerted millisecond timescale dynamics in the intrinsically disordered carboxyl terminus of γ-tubulin induced by mutation of a conserved tyrosine residue.Protein Sci. 2018 Feb;27(2):531-545. doi: 10.1002/pro.3345. Epub 2017 Dec 15. Protein Sci. 2018. PMID: 29127738 Free PMC article.
-
γ-Tubulin complexes in microtubule nucleation and beyond.Mol Biol Cell. 2015 Sep 1;26(17):2957-62. doi: 10.1091/mbc.E14-11-1514. Mol Biol Cell. 2015. PMID: 26316498 Free PMC article.
-
The cyclin-dependent kinase Cdc28p regulates multiple aspects of Kar9p function in yeast.Mol Biol Cell. 2007 Apr;18(4):1187-202. doi: 10.1091/mbc.e06-04-0360. Epub 2007 Jan 24. Mol Biol Cell. 2007. PMID: 17251549 Free PMC article.
-
Effect of GFP tags on the localization of EB1 and EB1 fragments in vivo.Cytoskeleton (Hoboken). 2010 Jan;67(1):1-12. doi: 10.1002/cm.20409. Cytoskeleton (Hoboken). 2010. PMID: 19701929 Free PMC article.
References
-
- Aldaz H., Rice L. M., Stearns T., Agard D. A. Insights into microtubule nucleation from the crystal structure of human γ-tubulin. Nature. 2005;435:523–527. - PubMed
-
- Beach D. L., Thibodeaux J., Maddox P., Yeh E., Bloom K. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr. Biol. 2000;10:1497–1506. - PubMed
-
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

