Phylogenetic analyses of cyanobacterial genomes: quantification of horizontal gene transfer events
- PMID: 16899658
- PMCID: PMC1557764
- DOI: 10.1101/gr.5322306
Phylogenetic analyses of cyanobacterial genomes: quantification of horizontal gene transfer events
Abstract
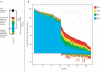
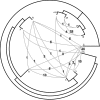
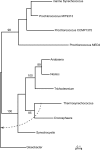
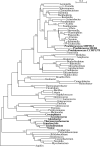
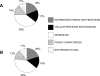
Using 1128 protein-coding gene families from 11 completely sequenced cyanobacterial genomes, we attempt to quantify horizontal gene transfer events within cyanobacteria, as well as between cyanobacteria and other phyla. A novel method of detecting and enumerating potential horizontal gene transfer events within a group of organisms based on analyses of "embedded quartets" allows us to identify phylogenetic signal consistent with a plurality of gene families, as well as to delineate cases of conflict to the plurality signal, which include horizontally transferred genes. To infer horizontal gene transfer events between cyanobacteria and other phyla, we added homologs from 168 available genomes. We screened phylogenetic trees reconstructed for each of these extended gene families for highly supported monophyly of cyanobacteria (or lack of it). Cyanobacterial genomes reveal a complex evolutionary history, which cannot be represented by a single strictly bifurcating tree for all genes or even most genes, although a single completely resolved phylogeny was recovered from the quartets' plurality signals. We find more conflicts within cyanobacteria than between cyanobacteria and other phyla. We also find that genes from all functional categories are subject to transfer. However, in interphylum as compared to intraphylum transfers, the proportion of metabolic (operational) gene transfers increases, while the proportion of informational gene transfers decreases.
Figures

References
-
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. - PMC - PubMed
-
- Andersson J.O., Sarchfield S.W., Roger A.J., Sjogren A.M., Davis L.A., Embley T.M., Sarchfield S.W., Roger A.J., Sjogren A.M., Davis L.A., Embley T.M., Roger A.J., Sjogren A.M., Davis L.A., Embley T.M., Sjogren A.M., Davis L.A., Embley T.M., Davis L.A., Embley T.M., Embley T.M. Gene transfers from Nanoarchaeota to an ancestor of diplomonads and parabasalids—Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Mol. Biol. Evol. 2005;22:85–90. - PubMed
-
- Bapteste E., Boucher Y., Leigh J., Doolittle W.F., Boucher Y., Leigh J., Doolittle W.F., Leigh J., Doolittle W.F., Doolittle W.F. Phylogenetic reconstruction and lateral gene transfer. Trends Microbiol. 2004;12:406–411. - PubMed
-
- Barker G.L., Handley B.A., Vacharapiyasophon P., Stevens J.R., Hayes P.K., Handley B.A., Vacharapiyasophon P., Stevens J.R., Hayes P.K., Vacharapiyasophon P., Stevens J.R., Hayes P.K., Stevens J.R., Hayes P.K., Hayes P.K. Allele-specific PCR shows that genetic exchange occurs among genetically diverse Nodularia (cyanobacteria) filaments in the Baltic Sea. Microbiol. 2000;146:2865–2875. - PubMed
-
- Baum B. Combining trees as a way of combining data sets for phylogenetic inference, and the desirability of combining gene trees. Taxon. 1992;41:3–10.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
