Calcium activation of the LMO4 transcription complex and its role in the patterning of thalamocortical connections
- PMID: 16899735
- PMCID: PMC6673794
- DOI: 10.1523/JNEUROSCI.0618-06.2006
Calcium activation of the LMO4 transcription complex and its role in the patterning of thalamocortical connections
Abstract
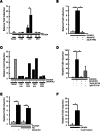
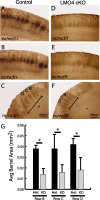
Lasting changes in neuronal connectivity require calcium-dependent gene expression. Here we report the identification of LIM domain-only 4 (LMO4) as a mediator of calcium-dependent transcription in cortical neurons. Calcium influx via voltage-sensitive calcium channels and NMDA receptors contributes to synaptically induced LMO4-mediated transactivation. LMO4-mediated transcription is dependent on signaling via calcium/calmodulin-dependent protein (CaM) kinase IV and microtubule-associated protein (MAP) kinase downstream of synaptic stimulation. Coimmunoprecipitation experiments indicate that LMO4 can form a complex with cAMP response element-binding protein (CREB) and can interact with cofactor of LIM homeodomain protein 1 (CLIM1) and CLIM2. To evaluate the role of LMO4 in vivo, we examined the consequences of conditional loss of lmo4 in the forebrain, using the Cre-Lox gene-targeting strategy. The organization of the barrel field in somatosensory cortex is disrupted in mice in which lmo4 is deleted conditionally in the cortex. Specifically, in contrast to controls, thalamocortical afferents in conditional lmo4 null mice fail to segregate into distinct barrel-specific domains. These observations identify LMO4 as a calcium-dependent transactivator that plays a key role in patterning thalamocortical connections during development.
Figures






References
-
- Ahn S, Riccio A, Ginty DD (2000). Spatial considerations for stimulus-dependent transcription in neurons. Annu Rev Physiol 62:803–823. - PubMed
-
- Aizawa H, Hu SC, Bobb K, Balakrishnan K, Ince G, Gurevich I, Cowan M, Ghosh A (2004). Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science 303:197–202. - PubMed
-
- Bading H, Ginty DD, Greenberg ME (1993). Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science 260:181–186. - PubMed
-
- Bulchand S, Subramanian L, Tole S (2003). Dynamic spatiotemporal expression of LIM genes and cofactors in the embryonic and postnatal cerebral cortex. Dev Dyn 226:460–469. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
